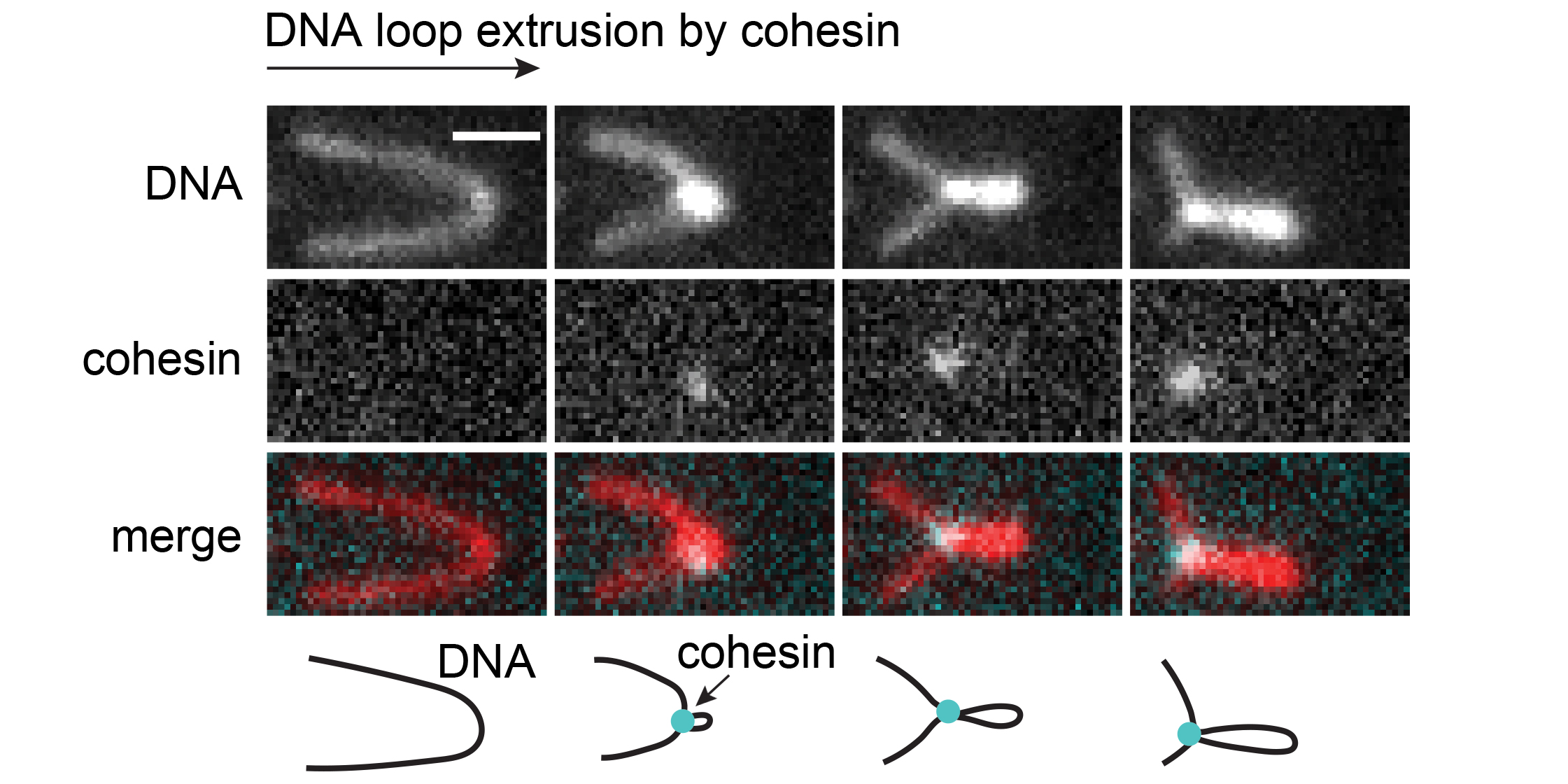
A research project (LS19-029) funded by the Vienna Science and Technology Fund (WWTF)

Eukaryotic genomes are folded into loops and topologically associating domains (TADs), which contribute to chromatin structure, gene regulation and recombination. These long-range interactions depend on cohesin, an ATPase complex first identified for its role in sister chromatid cohesion. Whereas cohesin is thought to mediate cohesion as a passive topological linker, it has been proposed that cohesin actively forms loops by an unknown extrusion mechanism. Consistent with the latter hypothesis we and others have recently found that cohesin is able to extrude DNA into loops in a process that depends on the protein NIPBL and on ATP hydrolysis by cohesin.
The aim of this project is to provide insight into the mechanism of DNA loop extrusion mediated by cohesin-NIPBL. For this purpose, we will image this process at multiple scales of spatial and temporal resolution, using a combination of high-speed atomic force microscopy, Förster resonance energy transfer and “time-resolved” cryo-electron microscopy. The function of cohesin movements visualized by these techniques will be tested by mutagenesis and confocal imaging in cells. This multimodal imaging approach will reveal how cohesin folds DNA, which will be key for understanding genome organization, regulation and function.
Project number LS19-029
Principal Investigator and Coordinator:
Jan-Michael Peters (Research Institute of Molecular Pathology, Vienna)
Co-Principal Investigators/Core Team Members:
Anton Golobordko (IMBA, Austrian Academy of Sciences, Vienna)
David Haselbach (Research Institute of Molecular Pathology, Vienna)
Peter Hinterdorfer (Johannes Kepler Universität Linz)
Further reading
- Davidson, I.F., Bauer, B., Goetz, D., Tang, W., Wutz, G. and Peters, J.-M. (2019). DNA loop extrusion by human cohesin. Science 366, 1388-1345.
- Davidson, I.F. and Peters, J.-M. (2021). Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol., doi: 10.1038/s41580-021-00349-7. Online ahead of print.
Links
https://www.imp.ac.at/groups/jan-michael-peters/
https://www.imp.ac.at/groups/david-haselbach/
https://www.oeaw.ac.at/imba/research/anton-goloborodko
https://www.jku.at/institut-fuer-biophysik/ueber-uns/team/angewandte-experimentelle-biophysik/