Watching molecular machines in action
Recently, single-particle cryo-EM has evolved to become one of the main methods of structural biology. While the technique was only available to visualise low-resolution “blobs” a few years ago, it is now capable of yielding high-resolution structures in a routine fashion.
To achieve this, the molecules are shock-frozen in a thin layer of ice and imaged in a transmission electron microscope (TEM), resulting in 2D projections of individual molecules. Computational analysis of several hundred thousand of these images gives rise to a 3D structure. While such structures can offer detailed insights into the function of a given macromolecular complex, there could be more information to find in the data.

In contrast to the other main structural techniques, the signals from each individual molecule can easily be distinguished in the TEM images. With these signals, it is possible to apply sophisticated sorting techniques to calculate an ensemble of different conformations that the molecular machine adopts simultaneously. This enables us to create movies of macromolecular machines in action. In addition, we can use statistical thermodynamics approaches to extract the thermodynamic and kinetic properties of the system.
The underlying experimental and computational approaches are still sparse and incomplete, however. Their completion is one of the Haselbach lab's main task. We use single molecule techniques to complement sparse kinetic data extracted through cryo-EM.
Malfunctioning machines
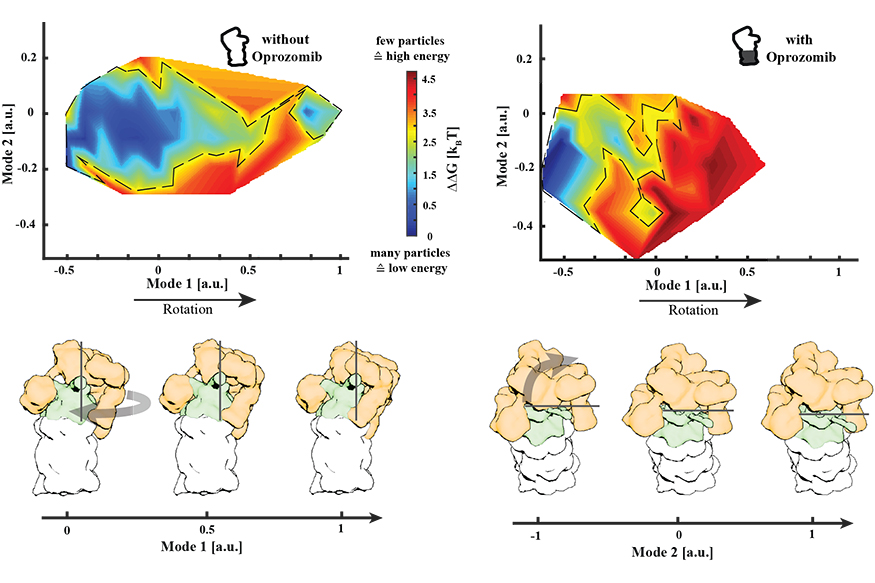
Despite their intricate design, molecular machines can fail. Using the biophysical methods described, we are endeavouring to understand the molecular response of molecular machines on stress and disease. In our initial study, we focused on the molecular adaption of the human 26S proteasome towards chemical inhibitors. Proteasomal degradation is a multi-step process involving substrate recognition, de-ubiquitination and unfolding carried out by the regulatory particle, as well as proteolysis performed by the core structure. Even though it has been well documented that the proteolytic activity of the core is shut down by using covalent inhibitors, the effect on the holoenzyme was unknown.
We identified a major allosteric effect that translates the binding of a single inhibitor to the proteolytic site to a restriction of the conformational space of the regulatory particle that is more than 15 nanometres away. We hypothesise that this can be sensed by different rescue factors that bind to the proteasome, resulting in a release of the applied stress.
Although the use of chemical inhibitors is clinically relevant, it is an artificial set-up. Our future aim is to understand natural causes of malfunctioning macromolecular machines and the processes involved in the repair of malfunctioning machines. We continue to focus on the 26S proteasome.
Here especially, the unfolding and translocation of the substrate into the proteolytic chamber is prone to error. In extreme cases, the entrance to the 20S proteasome is clogged by a substrate and degradation can no longer take place. A few prominent examples of proteasome stalling include the surface antigen of the Eppstein-Barr virus and the amyloid-forming proteins huntingtin or α-synuclein, which have the potential to clog the proteasome directly. Moreover, more general physiological situations such as oxidative stress, heat stress and arsenide intoxication can lead to a failure of the proteasome.
Due to its involvement in many processes, failure of the proteasome can lead to fatal consequences for the cell and even the organism. Several rescue mechanisms have been identified. In most cases, external protein factors bind to the periphery of the complex and change the degradation rate. However, in all cases is poorly understood how this is achieved mechanistically. We therefore aim to use our biophysical method set to identify the mechanisms that rescue stalled molecular machines.
Selected Publications
-
Bodrug, T., Welsh, KA., Bolhuis, DL., Paulаkonis, E., Martinez-Chacin, RC., Liu, B., Pinkin, N., Bonacci, T., Cui, L., Xu, P., Roscow, O., Amann, SJ., Grishkovskaya, I., Emanuele, MJ., Harrison, JS., Steimel, JP., Hahn, KM., Zhang, W., Zhong, ED., Haselbach, D., Brown, NG. (2023) Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C). Nat Struct Mol Biol. 30(11):1663-1674
-
Hodáková, Z., Grishkovskaya, I., Brunner, HL., Bolhuis, DL., Belačić, K., Schleiffer, A., Kotisch, H., Brown, NG., Haselbach, D. (2023) Cryo-EM structure of the chain-elongating E3 ubiquitin ligase UBR5. EMBO J. 42(16):e113348
-
Leesch, F., Lorenzo-Orts, L., Pribitzer, C., Grishkovskaya, I., Roehsner, J., Chugunova, A., Matzinger, M., Roitinger, E., Belačić, K., Kandolf, S., Lin, TY., Mechtler, K., Meinhart, A., Haselbach, D., Pauli, A. (2023) A molecular network of conserved factors keeps ribosomes dormant in the egg. Nature. 613(7945):712-720
-
de Almeida, M., Hinterndorfer, M., Brunner, H., Grishkovskaya, I., Singh, K., Schleiffer, A., Jude, J., Deswal, S., Kalis, R., Vunjak, M., Lendl, T., Imre, R., Roitinger, E., Neumann, T., Kandolf, S., Schutzbier, M., Mechtler, K., Versteeg, GA., Haselbach, D., Zuber, J. (2021)
AKIRIN2 controls the nuclear import of proteasomes in vertebrates. Nature. 599(7885):491-496 - Haselbach D, Komarov I, Agafonov DE, Hartmuth K, Graf B, Dybkov O, Urlaub H, Kastner B, Luehrmann R, Stark H (2018). Structure and Conformational Dynamics of the Human Spliceosomal Bact Complex.
Cell 172, 1–11, doi: 10.1016/j.cell.2018.01.010
Join us
- Master students and Post-docs: Contact David Haselbach with a letter of intent detailing why you want to join the lab.
- PhD students: Calls open 1 March and 1 September, apply here:
Vienna BioCenter PhD Program





