Mitosis and chromosome biology
How is sister chromatid cohesion established and maintained?
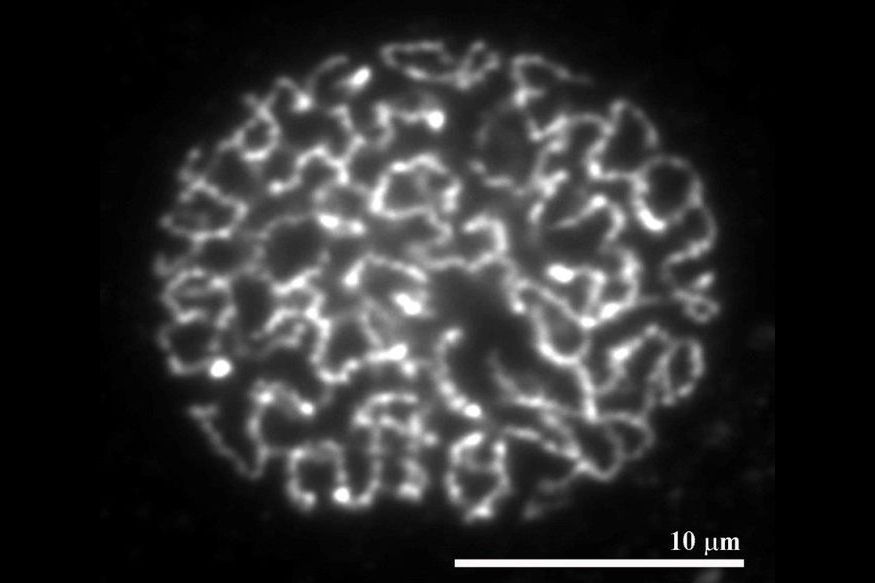
Numerous sites in the genome are bound by cohesin complexes. During DNA replication, these complexes establish physical connections between the newly synthesized sister chromatids. The resulting cohesion is known to be essential for chromosome segregation and DNA damage repair, but how cohesion is established and maintained is poorly understood. Our work indicates that cohesin is converted into a ‘cohesive’ form by the protein Sororin which: associates with cohesin during DNA replication; inhibits the cohesin release factor, Wapl; thus stabilizes cohesin on DNA; and enables cohesin to maintain cohesion. Our current work aims at understanding how Wapl releases cohesin from DNA, how Sororin prevents this, and how these proteins are regulated in mammalian oocytes where cohesion has to be maintained for long periods of time. In humans this is required for many years.
How does cohesin control chromatin structure and gene regulation?
Although cohesin can establish cohesion only during DNA replication, cohesin also associates with unreplicated DNA in the G1 phase and in post-mitotic cells. In these situations cohesin is not stabilized on DNA by Sororin, but can instead be released again by Wapl. We suspect that cohesin binds to unreplicated DNA in a dynamic manner because it also has important functions in structuring chromatin and gene regulation. Supporting this hypothesis, we found that: cohesin co-localizes with the transcriptional insulator protein, CTCF, in mammalian genomes; is required together with CTCF for gene regulation; and causes major changes in chromatin compaction when it is stabilized on DNA by the depletion of Wapl. In the future, we aim to understand how cohesin controls chromatin structure and gene regulation, and how Wapl might regulate these functions.
How is sister chromatid cohesion dissolved during mitosis?
Sister chromatid separation in anaphase depends on the removal of cohesin from chromosomes. This is mediated by Wapl which is activated in prophase and releases cohesin from chromosome arms, whereas centromeric cohesin is only removed from chromosomes in metaphase by the protease, separase. We recently discovered that the Wapl-dependent prophase pathway of cohesin release is important for complete separation of chromosome arms in anaphase, and simultaneously protects most cohesin complexes from cleavage by the protease, separase. This protection might be important for the next cell cycle, in which intact cohesin complexes are needed to re-establish a proper chromatin structure and gene expression programs.
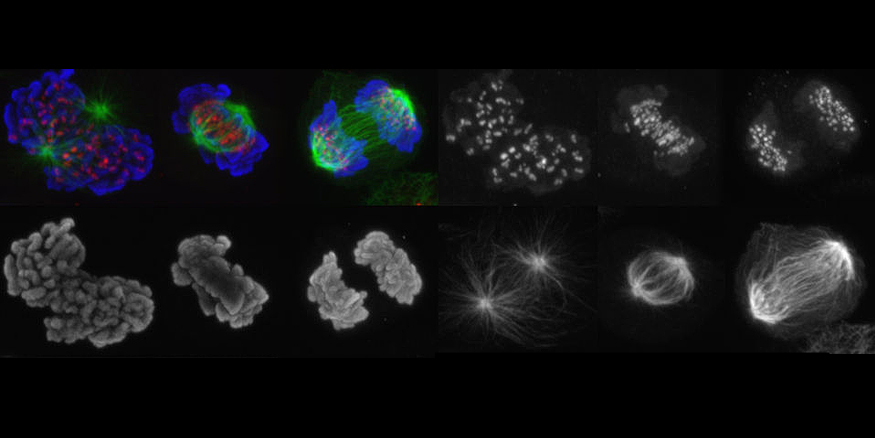
How does the APC/C initiate anaphase?
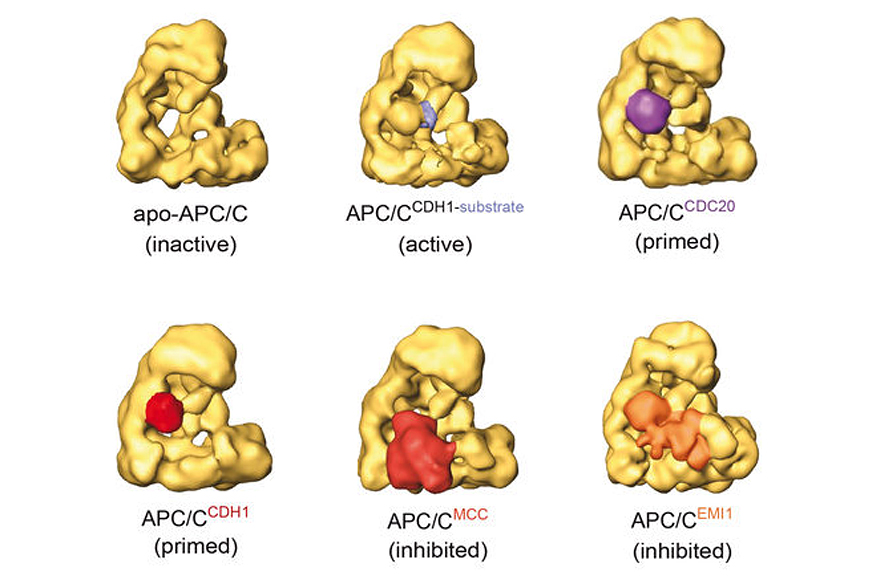
In metaphase, when all chromosomes have been bioriented, the anaphase-promoting complex/cyclosome (APC/C) is activated. The APC/C is a 1.5 MDa complex which assembles ubiquitin chains on securin and cyclin B. The subsequent destruction of these proteins by the 26S proteasome permits activation of separase, cleavage of centromeric cohesin, and sister chromatid separation. Until chromosome biorientation is complete, the APC/C is inhibited by the spindle assembly checkpoint (SAC).
The SAC ensures that sister chromatids are only separated once chromosomes have been attached to both spindle poles. Despite the importance of the APC/C, it is poorly understood how this complex is inhibited by the SAC, how the inhibition is relieved in metaphase, and how active APC/C recruits and ubiquitylates its substrates. We are using biochemical assays and electron microscopic analyses of the APC/C in different functional states to address these questions.
MitoCheck / MitoSys
Although mitosis has been studied for more than a century, our molecular understanding of this complex process is far from complete. From 2004 to 2009, the MitoCheck consortium, funded by the European Union, developed and applied genomic and proteomic approaches to study mitosis.
The consortium used RNA interference screens to identify proteins required for mitosis in human cells, tagging of genes in bacterial artificial chromosomes (BACs) to enable intracellular localization and affinity purification of these proteins, and mass spectrometry to identify protein complexes and mitosis-specific phosphorylation sites on these.
This work led to the identification of about 100 human protein complexes, many of which had been previously unknown or poorly characterized. Importantly, the approaches developed by MitoCheck will be generally applicable to high throughput analyses of other processes in mammalian cells. In a new project funded by the European Union, known as MitoSys (2010 to 2015), we are developing quantitative assays for mitosis.
Mitosys Documentary

The documentary, Meetings of Minds, allows scientists to speak for themselves in conversation with artists. Four artist/scientist pairs have been selected to approach the subject of mitosis from different perspectives:
Ways of Saying
Shobana Jeyasingh / Kim Nasmyth
Ways of Seeing
Rob Kesseler / Melina Schuh
Ways of Being
Ackroyd & Harvey / Jan-Michael Peters
Ways of Growing
Lucy & Jorges Orta / Tony Hyman
Selected Publications
-
Davidson, IF., Barth, R., Zaczek, M., van der Torre, J., Tang, W., Nagasaka, K., Janissen, R., Kerssemakers, J., Wutz, G., Dekker, C., Peters, JM. (2023). CTCF is a DNA-tension-dependent barrier to cohesin-mediated loop extrusion. Nature. 616(7958):822-827
-
Panarotto, M., Davidson, I.F., Schleiffer, A. and Peters, J.-M. (2022). Cornelia de Lange Syndrome mutations in NIPBL can impair cohesin-mediated DNA loop extrusion. Proc Natl Acad Sci U S A, 119, e2201029119.
- Bauer, B., Davidson, I.F., Canena, D., Wutz, G., Tang, W., Litos, G., Horn, S., Hinterdorfer, P. and Peters, J.-M. (2021). Cohesin mediates DNA loop extrusion by a “swing and clamp” mechanism. Cell 184, 5448-5464.
- Davidson, I.F., Bauer, B., Goetz, D., Tang, W., Wutz, G. and Peters, J.-M. (2019). DNA loop extrusion by human cohesin. Science 366, 1388-1345.
- Busslinger, G.A., Stocsits, R.R.,van der Lelij, P., Axelsson, E., Tedeschi, A., Galjart, N. and Peters, J.-M. (2017). Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature, 544, 503-507.
Join us
- Master students and Post-docs: Contact Jan-Michael Peters with a letter of intent detailing why you want to join the lab.
- PhD students: Calls open 1 March and 1 September, apply here:
Vienna BioCenter PhD Program
Research questions and approaches
We use a YouTube plugin to display social media content on this page, which places requests to YouTube servers. These requests make your IP address visible to YouTube, who may use it in accordance with their data privacy policy. Please agree to make the YouTube video visible. You can find more information in our privacy settings.
AcceptGrants

Marie Curie ITN: 101072505 CohesiNet