Inside the cell’s recycling hub: unveiling the architecture of UBR4
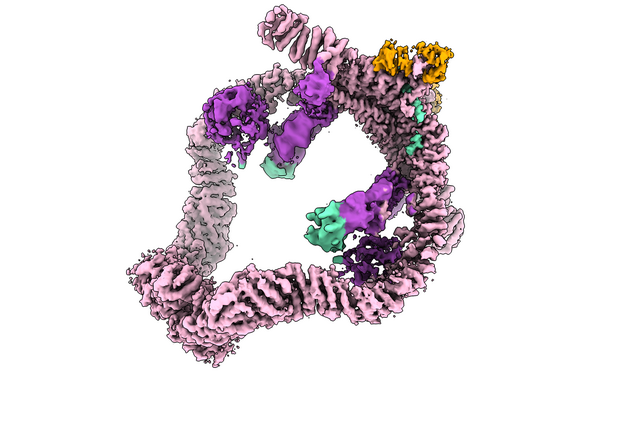
Researchers from the lab of Tim Clausen and collaborators have unveiled the three-dimensional structure of UBR4, a giant protein complex that safeguards cells by targeting defective proteins for destruction—offering fresh insights into health, disease, and potential cancer therapies. Their findings were now published in the journal Science.
Inside every cell, thousands of different proteins work in concert, each performing unique, essential functions to keep the cell fit and thriving. This is a delicate choreography, where functional proteins must be maintained in peak condition, while defective proteins are swiftly removed before they can cause harm. To ensure this, cells employ protein quality control systems—molecular guardians that detect misfolded, damaged, or misplaced proteins and mark them for destruction using a chemical “tag” called ubiquitin.
At the heart of one of these clean-up crews is UBR4, a protein tasked with identifying proteins already marked with ubiquitin and further processing them so the cell’s waste-disposal machinery can degrade them. To do this, UBR4 relies on its cofactors KCMF1 and CALM1, forming a large protein complex that acts as a hub for selective degradation of defective proteins. In doing so, UBR4 acts much like a recycling foreman—sorting the defective parts and ensuring they are sent down the correct conveyor belt for dismantling. However, despite its remarkable importance, the three-dimensional structure of UBR4’s complex has remained poorly understood, limiting our understanding of how URB4 performs its essential function.
Now, a team led by Daniel Grabarczyk, a research associate in Tim Clausen’s lab at the Research Institute of Molecular Pathology (IMP), together with collaborators, has captured the first snapshot of UBR4 working together with its cofactors, providing new insights into the structure and function of this complex. The effort drew from scientists at a range of institutions: the Max Perutz Labs, the University of Vienna, Medical University of Vienna, Korea University, and the MRC Laboratory of Molecular Biology (UK).
Cryo-EM reveals a ring-shaped degradation hub
To determine the URB4 complex’s structure, the scientists employed advanced cryo-electron microscopy (cryo-EM) techniques. They discovered that UBR4 and its cofactors form a giant, intricate, ring-shaped protein complex that serves as a hub where faulty proteins are brought in, inspected and processed for final disposal.
This structural view not only shows the overall architecture of the URB4 complex, but also the inner workings of its members. The team discovered that UBR4 serves as the main catalytic engine, extending ubiquitin chains to guide proteins to degradation. Instead, its partner KCMF1, once thought to carry out this catalytic role, turned out to work as an adapter, helping the complex selectively recognise “address labels” in marked proteins to help select those that should be degraded.
“Our approach also allowed us to observe different stages of the URB4 complex’s function, revealing how it adapts to handle a wide variety of defective proteins,” Tim Clausen comments: “Thanks to our great collaborators Hyun Kyu Song at Korea University, Ramanujan Hegde at the MRC LMB, and other colleagues, we were able to extend our study to reveal the biophysical and structural principles of substrate selection, providing a rationale for how UBR4 cooperates with other quality-control systems in the cell.”
By comparing UBR4 complexes from humans, the roundworm Caenorhabditis elegans, and the plant Arabidopsis thaliana, the team found that while the overall blueprint of the URB4 complex is conserved across eukaryotes, evolutionary tweaks have optimized the machinery for different cellular environments—whether in plant leaves or human neurons.
A potential Achilles’ heel for cancer cells
The team’s findings shed new light on how cells clean up defective proteins, a process not only relevant to keeping cells healthy, but also to cancer cell survival. Cancer cells depend on UBR4 to maintain their own internal balance, especially in tumours with high levels of chromosome instability. Understanding the molecular details of how UBR4 and its partners work opens a potential new route for drug development: designing molecules that selectively disrupt this complex could be a promising therapeutic strategy.
Original publication
Grabarczyk D., Ehrmann J., Murphy P., Yang W., Kurzbauer R., Bell L., Deszcz L., Neuhold J., Schleiffer A., Shulkina A., Lee J., Shin KJ., Meinhart A., Versteeg G., Zavodszky E., Song H., Hegde R., Clausen T.: “Architecture of the UBR4 complex, a giant E4 ligase central to eukaryotic protein quality control”. Science. DOI: 10.1126/science.adv9309
About the IMP
The Research Institute of Molecular Pathology (IMP) in Vienna is a basic life science research institute largely sponsored by Boehringer Ingelheim. With over 200 scientists from 40 countries, the IMP is committed to scientific discovery of fundamental molecular and cellular mechanisms underlying complex biological phenomena. The IMP is part of the Vienna BioCenter, one of Europe’s most dynamic life science hubs with nearly 3,000 employees from 80 countries in seven research organisations, two universities, and more than 45 biotech companies. www.imp.ac.at, www.viennabiocenter.org