"Heart" of the spliceosome: putting together a molecular machine
Splicing is a fundamental process in the expression of our genetic information. It requires a complex molecular machinery, which needs to be made and recycled. Both of these processes are poorly understood, but scientists from the lab of Clemens Plaschka now added an important piece of the puzzle: they showed how the molecular "scaffolding" of the splicing machinery transforms through four stages to return to an early state. Their findings were published in the journal Nature Structural and Molecular Biology.
RNA splicing is a process in organisms with a defined cell nucleus, such as animals and plants, which turns newly formed RNA into mature messenger RNA. The messenger RNA then exits the nucleus, carrying instructions for the assembly of proteins to the organelles where the proteins are built. In short: RNA splicing is a fundamental and vital step in all cells of most living beings. Scientists increasingly understand the act of splicing, but they know relatively little about what follows it – such as the recycling of the components that form the splicing machinery.
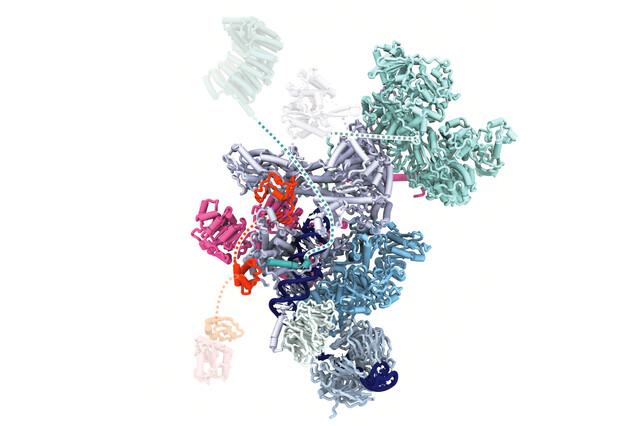
"The spliceosome is a large and complex structure that forms anew each time it processes RNA. This happens through the re-arrangement of five central parts of the spliceosome, the snRNPs," says Daria Riabov Bassat, postdoc and co-first author of the study. Studying these parts is challenging and requires a range of tools first to isolate the spliceosome at the right stage and then to analyse its structure at the molecular scale using cryo-electron microscopy. The Plaschka lab has great expertise in both these fields, and so Riabov Bassat, her co-first author Supapat Visanpattanasin, and others eventually succeeded by collecting vast amounts of structural data.
"Using human samples, we can now show how the ‘heart’ of the spliceosome is made and recycled in unprecedented resolution," says Daria Riabov. This ‘heart’, called the U5 snRNP, plays an important role in forming the scaffold for other components of the spliceosome. "We describe four cryo-EM structures that show how this heart is recognised after splicing, how its key domains are remodelled, and how the binding of additional elements is controlled, to make and recycle the U5 snRNP for a new round of splicing."
The structures and their function in the management of the spliceosome are an important contribution to the general research question of the Plaschka lab – of how molecular machines prepare messenger RNA for gene expression.
"Our results provide a new conceptual model for how spliceosome subunits are formed and recycled: specific chaperones would organise snRNP domains, guide the binding of additional subunits, and coordinate these rearrangements with the dissociation of the chaperones," says Clemens Plaschka.
The discovery opens new angles for related research. With the role of U5 snRNP as a kind of molecular scaffold firmly established, the scientists will now seek to learn more about other structures that rely on it.
Original Publication
Daria Riabov Bassat, Supapat Visanpattanasin, Matthias K. Vorländer, Laura Fin, Alexander W. Phillips, and Clemens Plaschka (2024): “Late biogenesis and recycling of the human spliceosomal U5 snRNP”. Nature Structural and Molecular Biology. DOI: 10.1038/s41594-024-01243-4
Related publication
This study was published alongside with related work by scientists from the lab of Wojtek Galej at EMBL Grenoble on "Structure of the human 20S U5 snRNP". DOI: 10.1038/s41594-024-01250-5
Further Reading