DNA loop formation linked to Cornelia de Lange Syndrome
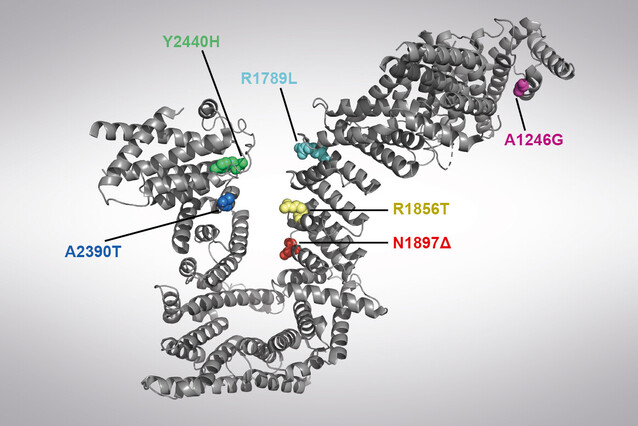
Healthy cells package their DNA with the help of proteins that fold it into loops. Scientists from the lab of Jan-Michael Peters discovered that the formation of DNA loops can be impaired by mutations found in Cornelia de Lange Syndrome (CdLS), a neurodevelopmental disorder. The discovery may identify a main cause of the disorder and was now published in the journal PNAS.
Cornelia de Lange Syndrome (CdLS) is a developmental disorder, characterised by abnormal physical, cognitive, and behavioural traits. The condition was named after the Dutch paediatrician Cornelia de Lange, who first published on it in 1933. CdLS is thought to arise from developmental genes being activated - or expressed - at the wrong time and intensity, but the exact molecular cause is still unclear.
There are several cellular mechanisms known to control the proper activation of genes, including the formation of DNA loops. Most of these loops are formed by cohesin, a ring-shaped protein complex of which the protein NIPBL is a regulatory subunit. Strikingly, NIPBL mutations are detected in about 70 percent of persons suffering from Cornelia de Lange Syndrome, and gene expression changes are also associated with the condition.
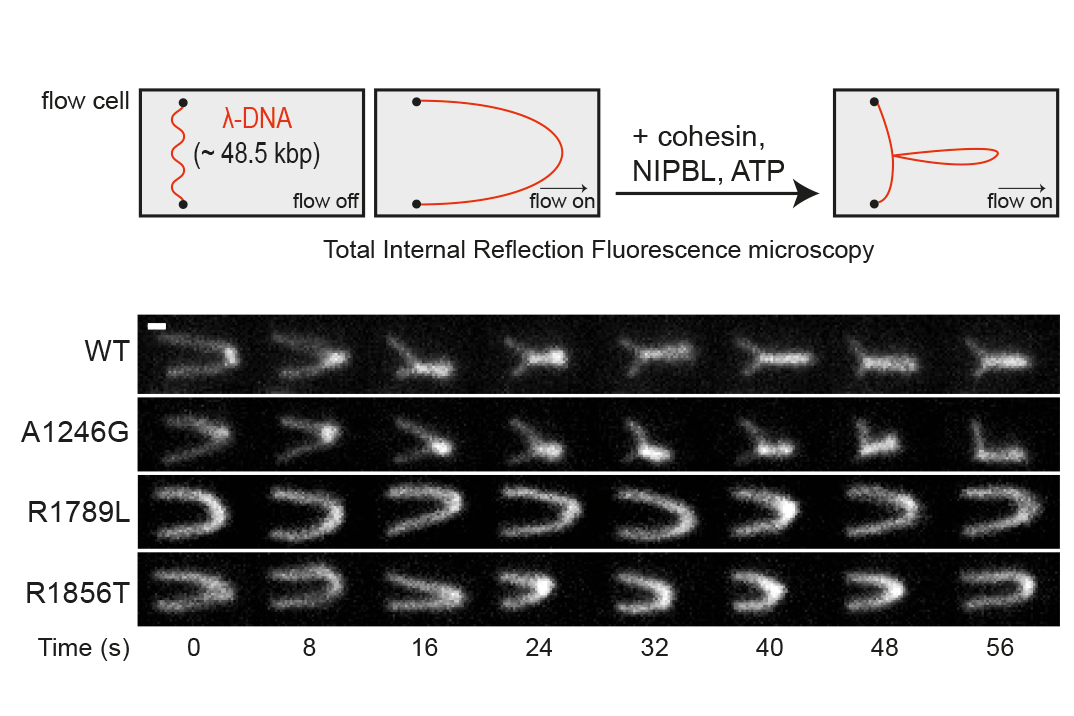
Scientists from the lab of Jan-Michael Peters thus wondered whether mutations in NIPBL cause defects in making DNA loops. Using a method developed by IMP scientist Iain Davidson a few years ago, they observed the extrusion of DNA loops by cohesin in the presence of NIPBL - at single-molecule scale and in real time. They found that six NIPBL mutations indeed impair the loop extrusion activity of cohesin. The resulting study, now published in the journal PNAS, links these mutations to a mechanism underlying CdLS.
“Defects in cohesin-mediated loop extrusion could be one cause, or perhaps even the root cause, of human developmental diseases, including Cornelia de Lange Syndrome,” says Jan-Michael Peters. In cells, defects making chromatin loops would result in the mis-expressions of developmental genes. “Unfortunately, this insight does not open any new treatment options for CdLS patients. Still, any information on the causes of this disease might become helpful in the future.” Beyond the disease mechanism dimension, the study contributes to a broader debate in the cohesin field on whether DNA loop extrusion is required for the regulation of gene expression.

“Even small alterations in gene regulation can have a strong effect on human development from the earliest stages,” says Iain Davidson. “Defects in DNA loop formation and the resulting changes of gene expression could explain the severe abnormalities in patients with Cornelia de Lange Syndrome.”
“Cornelia de Lange Syndrome affects one of 10,000 to 30,000 newborns and is considered a rare disease. However, it shares symptoms with many other neurodevelopmental disorders,” says Melanie Panarotto. “This is why our findings might also give insights into the causes of those other related disorders.”
Original Publication
Melanie Panarotto, Iain F. Davidson, Gabriele Litos, Alexander Schleiffer, and Jan-Michael Peters (2022): “Cornelia de Lange Syndrome mutations in NIPBL can impair cohesin-mediated DNA loop extrusion”. PNAS. https://doi.org/10.1073/pnas.2201029119