A tale of DNA loops: how distant DNA segments come together to make different types of antibodies

To fight off the vast array of pathogens we encounter every day, our immune system has come up with genetic mechanisms to diversify pathogen-detecting antibodies. Antibodies can be grouped in various classes with different functions: for each class to be produced, the DNA segments that encode them must be rearranged through ‘Class Switch Recombination’. In a new study now published in the journal Molecular Cell, researchers in the lab of Rushad Pavri use cutting-edge methods to investigate this process and revise a long-standing model.
Our immune system detects and recognises millions of different foreign particles – or antigens –which circulate in our bodies in the early stages of an infection or after vaccination. To neutralise antigens, B cells produce a wide diversity of antibodies: these Y-shaped proteins carry highly variable antigen-binding sites (the Y’s two tips) joined to one of several constant regions (simply put, the Y’s trunk). The constant regions define an antibody’s class and confer different functions in the immune system aimed at destroying the antigen and clearing it from circulation.
Evolution has found an original way of generating this staggering diversity of antibodies: DNA mutations. While changes in our DNA are often associated with genetic diseases and cancer, they occur in B cells every day and allow us to thwart infectious microbes and obtain protective immunity after vaccination.
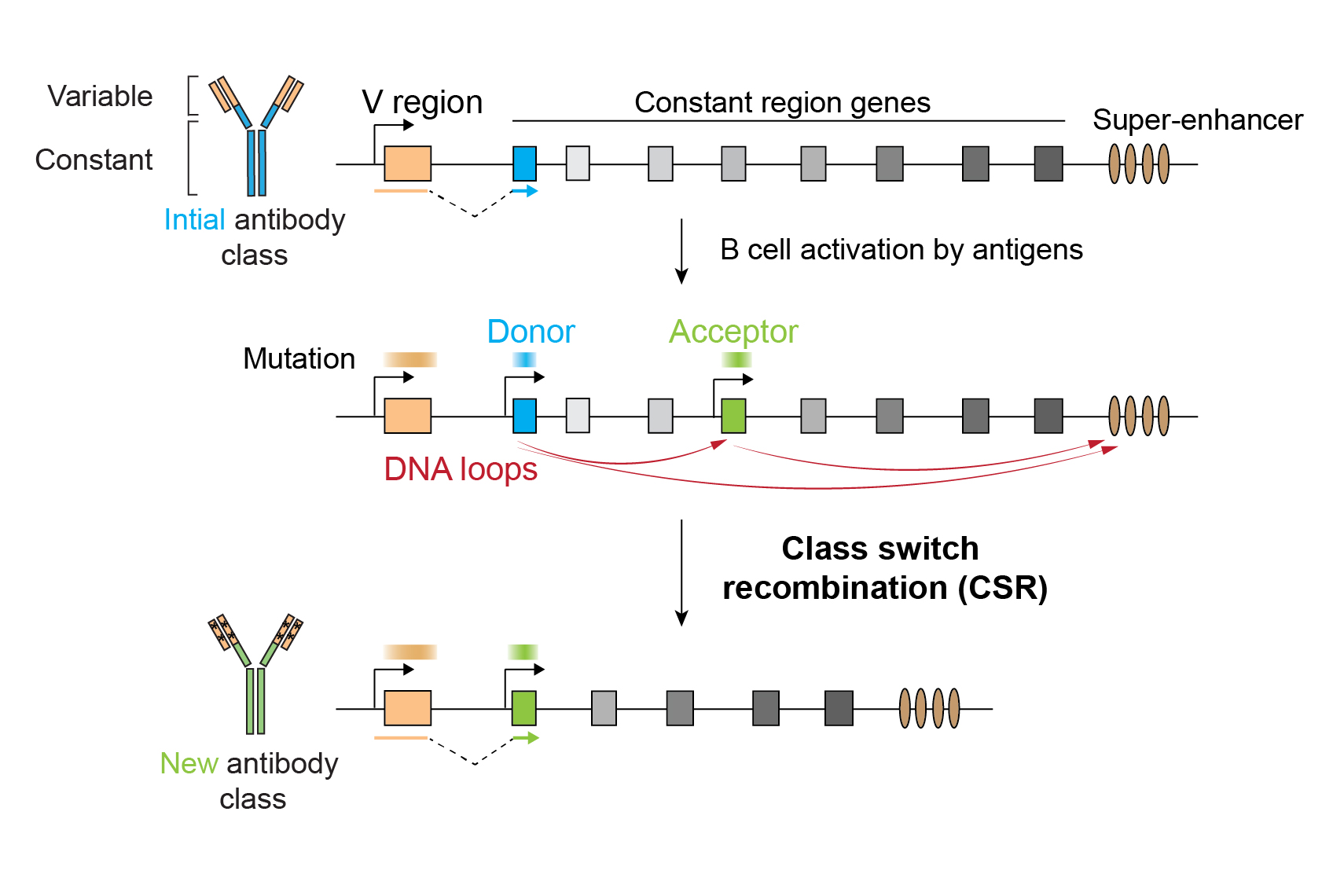
Class Switch Recombination (CSR) is a genetic mechanism that changes a B cell’s production of antibodies from one class to another. The DNA segment that is used to produce antibodies contains genes that encode the antigen-binding site, the variable region, followed by several possible constant regions. When antibody production begins, only the constant region directly adjacent to the variable region (called the ‘donor’) is used. Therefore, if a B cell is to produce another class of antibodies, the first region – or regions – has to be cut out to make way for another constant region (called the ‘acceptor’) (see figure below).
Mechanisms of Class Switch Recombination
To ensure proper CSR, DNA needs to be cut at the donor and acceptor constant regions, and the cut ends need to be glued together to produce an antibody containing the acceptor sequence. Transcription of the donor and acceptor sequences opens up the DNA to allow the cutting machinery to act. Both transcription and DNA cutting are fuelled with the help of a distant regulatory sequence or ‘super-enhancer’. Hence, the three participating DNA elements – the donor, acceptor and super-enhancer – must be brought close to one another for correct recombination. For this to occur, cohesin, a molecular motor that folds our two-meter-long genome to fit inside each of our cells, plays a crucial role. It binds DNA and reels it into a loop, such that two segments that were initially far apart come face to face and interact.
In the longstanding existing model of CSR, researchers have hypothesised that cohesin forms two DNA loops that bring the donor, the acceptor, and the super-enhancer sequences together, such that transcription and recombination can happen simultaneously in a three-way interaction (see figure below). However, three-way interactions have never been directly measured. Furthermore, prior studies did not have the resolution to determine which parts of the long donor, acceptor, and super-enhancer sequences make contact.
In a new study now published in the journal Molecular Cell, scientists in the lab of Rushad Pavri at the IMP address and refine this model, in collaboration with the lab of Anton Goloborodko at the neighbouring Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA).
Revising the older model
Julia Costea, co-first author and now a PhD student at the European Molecular Biology Laboratory, joined the IMP in 2019 to conduct her Master’s thesis research. “I came to the IMP at the right time for this project: a new method called ‘Tri-C’ had just been published that allowed us to directly measure multi-way interactions between DNA segments at high resolution. So far, we had only been able to visualise two-way interactions, and at a much lower resolution,” says Costea.
In their revised model, the researchers propose that, first, cohesin-mediated loops bring the super-enhancer into contact with the donor and acceptor sequences to initiate transcription. However, in contrast to the older model, they suggest that the three-way interaction between donor, acceptor, and super-enhancer is only transient, as it was barely detectable using Tri-C. The scientists also found that transcription created a physical boundary for cohesin in the donor and acceptor DNA: when new rounds of loop formation occur, cohesin molecules stop at the boundary such that acceptor and donor regions are now in close proximity. This barrier stabilises the interaction between donor and acceptor sequences and constitutes a new feature in the CSR mechanism (see figure below). While cohesin is known to halt at transcribed regions, this study provides the first piece of evidence that such genomic events can play an important role in key biological processes.
“I find it fascinating that several processes – cohesin folding DNA into loops, transcription stopping the folding at the right place and time – can drive a biological function that is so crucial for our immune system and our survival in a world full of potential pathogens,” says Ursula Schöberl, co-first author and postdoc.
“Our findings change the way we think about how DNA looping regulates Class Switch Recombination and give us insights beyond the field of immunology, touching upon genome architecture and gene regulation,” says Rushad Pavri. “This work results from the tight collaboration between the IMP and IMBA, and between bench scientists, computational biologists, and bioinformaticians. Having a host of experts in all fields of molecular biology at the Vienna BioCenter catalyses such interdisciplinary projects.”
Original publication
Julia Costea*, Ursula E. Schoeberl*, Daniel Malzl, Maximilian von der Linde, Johanna Fitz, Ankit Gupta, Marina Makharova, Anton Goloborodko and Rushad Pavri: “A de novo transcription-dependent TAD boundary underpins critical multiway interactions during antibody class switch recombination”. Molecular Cell (2023), DOI: 10.1016/j.molcel.2023.01.014.
*co-first authors
Further reading
Scientists uncover secret behind lymphoma-associated mutations