Artificial Intelligence cracks code of gene regulation
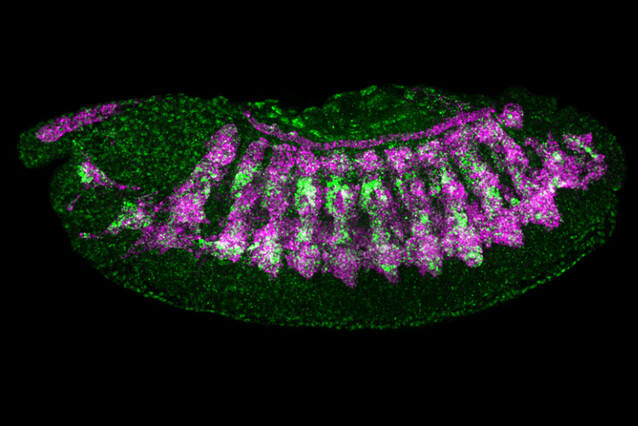
Organisms are built from thousands of different proteins, each of them encoded by a specific gene. For a cell type to acquire its unique identity, form, and function, it must activate genes through the expression of "enhancers". Scientists have long tried to crack the code by which enhancers operate. The labs of Alexander Stark at the IMP and Eileen Furlong at EMBL Heidelberg now harnessed genomics and artificial intelligence to solve this second "code of life". Their findings were published in the journal Nature.
Each healthy cell of a complex organism contains the exact same copy of the genome, which includes thousands of genes, the blueprints for building proteins. To form different cell types, tissues, and organs, additional mechanisms switch the expression of specific genes on and off with high precision.
‘Enhancers’ are genomic DNA segments that are key factors for switching on genes. The lab of IMP senior group leader Alexander Stark has made it its mission to crack the code that links an enhancer’s DNA sequence with its gene-regulatory function. While the first enhancers were discovered in the early 1980s, scientists developed methods to identify enhancers experimentally only in the past decade.
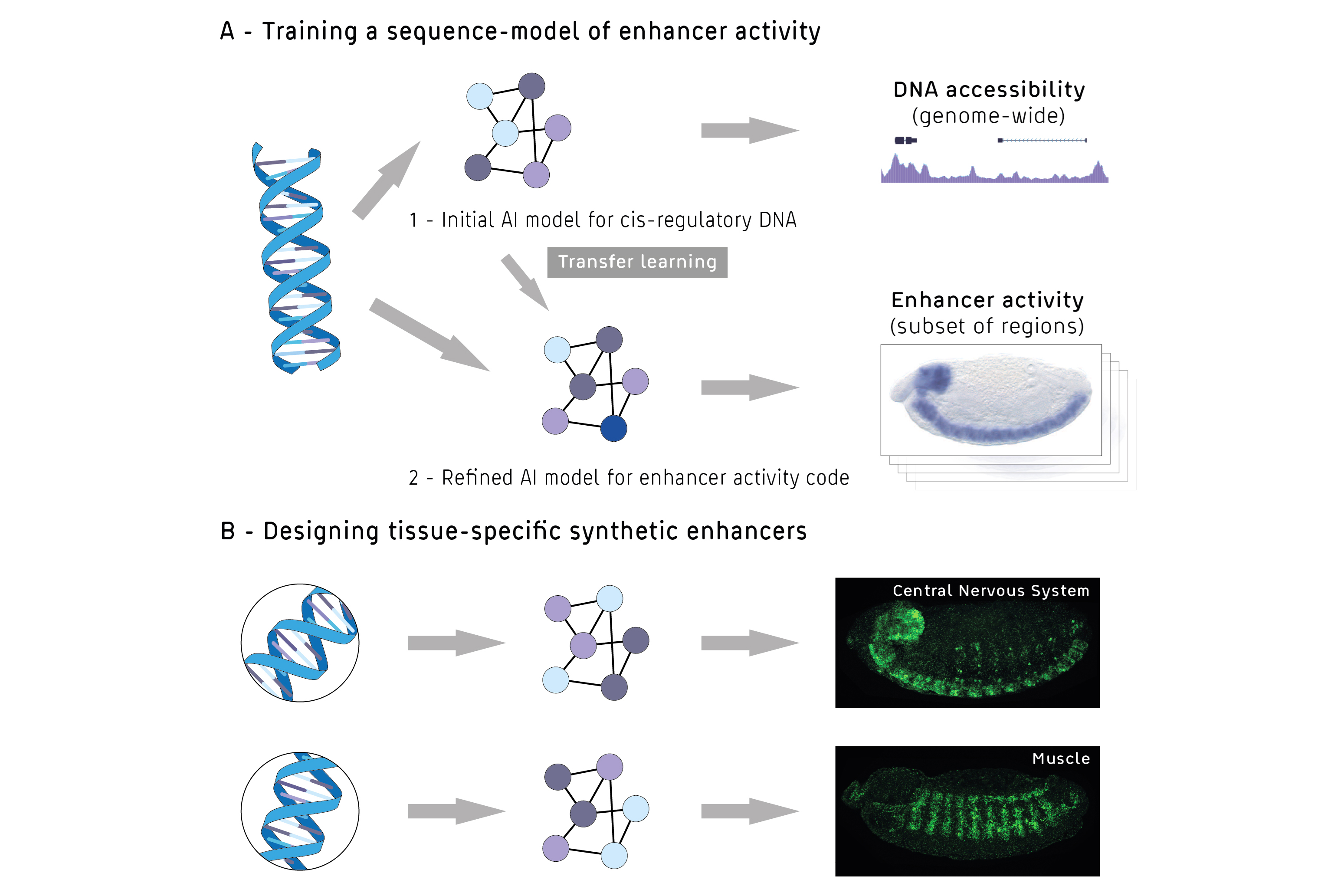
Building on this, the Stark lab and collaborators now aimed at three tasks that together form a long-held goal that seemed impossible to achieve: to predict the activity of enhancers from their DNA sequence; to predict consequences of mutations in enhancers; and to design enhancers from scratchfor specific tissues. In other words: to read, understand, and write the second genetic code, the code that underlies gene regulation.
An opportunity to crack this code emerged with recent advances in genomics and artificial intelligence. The scientists developed a powerful deep and transfer learning model, which they trained on large sets of data from previous studies in the fruit fly Drosophila melanogaster, a widely used model organism in developmental biology.
From the lab to AI and back
First, the model was trained on genome-wide DNA sequences and corresponding DNA accessibility data, a rather easily measured predictor of enhancers. This first model was then used to initialise the fine-tuning of a second model, which learned to directly link DNA sequences to specific enhancer activities.
"You could explain transfer learning as such: imagine you want to train a model to recognise cats in pictures, but you have only few cat pictures available. However, you do have many dog pictures. So you first train an AI model on dog pictures, and then fine-tune it in a second step to now recognise cats," says Alexander Stark.
With transfer learning, the model was able to predict enhancer activity for five types of tissues in fruit fly embryos: central nervous system, the sub-section of the brain, the epidermis, the gut, and muscle.
Building on this prediction, the scientists took their work back from the abstract world of big data and artificial intelligence and returned to the lab bench. Using well-established molecular biology tools, the scientists tested 40 synthetic enhancers that were designed computationallyin living fruit fly embryos. And indeed - the enhancers were active and drove gene expression in the targeted tissues.
Video: AI cracks code of gene regulation
"Being able to build synthetic enhancers with specific properties opens unprecedented opportunities for controlling the targeted expression of genes," says Bernardo de Almeida, first author of the study and recent graduate of the Vienna BioCenter PhD Program. Future applications could be in synthetic biology or gene therapy, where the precise design and manipulation of gene expression patterns is a prerequisite.
For Alexander Stark, however, the insight into a fundamental phenomenon of life is the most important aspect of the study: "About 60 years ago, scientists learned how the first genetic code works, how a molecular DNA blueprint can be translated into a protein," says Alexander Stark. "With the power of genomics and artificial intelligence, we have now managed to crack the second code of life – that of how gene activity is controlled. This study is a breakthrough and the peak of my research ever since I started my lab at the IMP in 2008."
Original Publication
Bernardo P. de Almeida, Christoph Schaub, Michaela Pagani, Stefano Secchia, Eileen E. M. Furlong, Alexander Stark. “Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo”. Nature (2023). DOI: 10.1038/s41586-023-06905-9
Related publication
This study was published alongside with related work by scientists from VIB-KU Leuven on ‘Cell type directed design of synthetic enhancers’. DOI: 10.1038/s41586-023-06936-2.
Further reading
Harnessing artificial intelligence to predict and control gene regulation (12 May 2022)
Decoding the grammar of gene regulation (20 March 2023)
Stark Lab: https://www.starklab.org/
Bernardo de Almeida:https://bernardo-de-almeida.github.io/
Eileen Furlong Lab: http://furlonglab.embl.de/
About the Vienna BioCenter PhD Program
Much of the work underlying this publication was done by a doctoral student of the Vienna BioCenter PhD Program. Are you interested in a world-class career in molecular biology? Find out more: https://training.vbc.ac.at/phd-program/
About the IMP at the Vienna BioCenter
The Research Institute of Molecular Pathology (IMP) in Vienna is a basic life science research institute largely sponsored by Boehringer Ingelheim. With over 200 scientists from 40 countries, the IMP is committed to scientific discovery of fundamental molecular and cellular mechanisms underlying complex biological phenomena. The IMP is part of the Vienna BioCenter, one of Europe’s most dynamic life science hubs with more than 3,000 people from over 80 countries in six research institutions, two universities, and 40 biotech companies. www.imp.ac.at,www.viennabiocenter.org