Luisa Cochella is an Adjunct Investigator at the IMP and Assistant Professor at Johns Hopkins University School of Medicine. You find more detailed information about her work and research group at: https://mbg.jhmi.edu/people/luisa-cochella-ph-d/
Transcriptional and post-transcriptional mechanisms of cell-type specification
Description of research activities
Cellular identities are defined by the expression of so-called terminal gene batteries that provide each cell type with its unique structural and functional properties. These defining gene batteries are in turn produced by the combinatorial action of gene expression regulators both at the transcriptional and at the post-transcriptional level. We aim to understand how the gene regulatory networks that control cell type differentiation are established during development and how they diversify both in development and through evolution. For this, we focus on two specific questions:
- What is the contribution of post-transcriptional regulators, such as miRNAs, to generating cellular complexity during development and evolution?
- How do early transcriptional events during development impact the final fate of a cell and translate into increased cellular diversity?
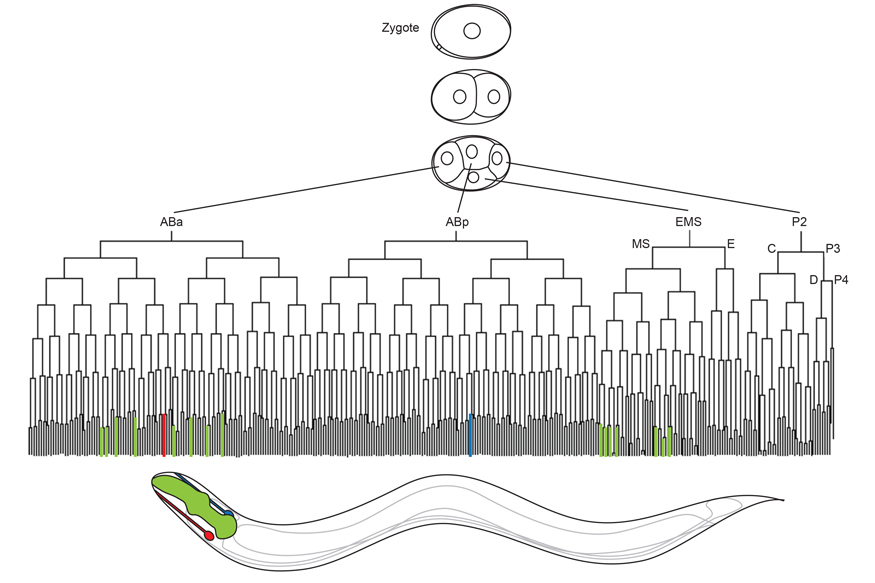
We address these questions using the nematode C. elegans as a model system as it has a well-defined anatomy with 959 somatic cells in the adult, which are produced by an invariant developmental lineage (Figure 1). This means that the developmental history of every cell can be followed from the zygote with certainty and shows how a relatively small number of cell divisions is sufficient to produce differentiated cells from the zygote. In addition, C. elegans is a powerful genetic model organism, amenable to classical approaches but also to more recent genome engineering methods with relative ease and speed. And finally, the output of genetic manipulations on cell type specification can be measured at the level of gene expression changes in individual cells as well as at the level of cellular function and organism physiology.
1. Understanding the role of post-transcriptional regulation by miRNAs in development
MicroRNAs are ideally suited to diversify genetic programs and contribute to the variety of cell types that constitute an adult organism. They are repressors of gene expression that have expanded during evolution, coinciding with the onset of multicellularity, and many of them display complex and dynamic spatio-temporal expression patterns. As such, miRNAs have the potential to modify pre-existing genetic networks to produce stable and heritable contributions to cell identities.
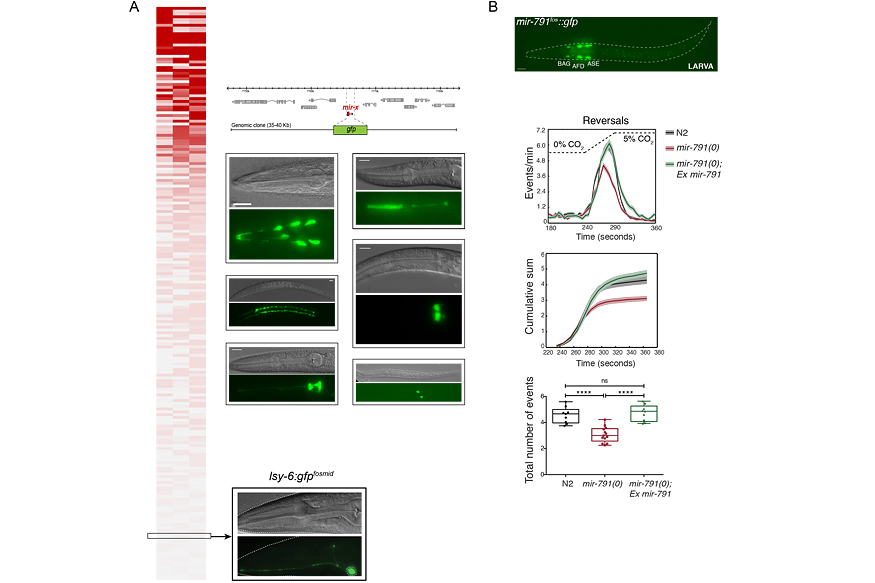
While miRNAs as a whole, as assessed by removal of the miRNA biogenesis machinery, are clearly important for development in animals and plants, the roles of specific miRNAs in development have been challenging to uncover. Systematic analysis of miRNA mutants in C. elegans failed to reveal obvious developmental defects for most miRNAs. Based on our miRNA expression analysis, we hypothesized that one of the main reasons for this is that most miRNAs are likely to be expressed with very high cellular specificity and therefore their functions may only be uncovered with very specific assays (Figure 2A). We have tested this hypothesis by studying the functions of specific miRNAs and in this way we have uncovered unknown functions for miRNAs in the production of specialized cells (Figure 2B).
In C. elegans up to ¾ of all miRNAs may be restricted to specific cell types. However, the remaining miRNAs are broadly and abundantly expressed in the embryo. We are currently exploring their contribution to development. Together we aim to provide a comprehensive framework for understanding the roles of miRNAs in development in C. elegans by providing a classification of miRNAs in two main groups: broadly expressed miRNAs with core developmental functions, and cell-type specific miRNAs with defined roles at more terminal stages of development. Our analysis of miRNA expression in other animal models suggests that a similar classification will apply as well.
A further advantage of working with C. elegans is that the genomes of a number of related nematode species have now been fully sequenced, and these species are being extensively characterized at various levels. This provides us with the possibility to compare gene regulatory networks that have acquired a miRNA component specifically in some of these species but not in others to test the contribution of miRNAs during evolution.
Conceptually similar mechanisms have been proposed during the specification of different lineages in other animals. Our goal is to use the worm to dissect one such mechanism, test the extent to which such a strategy is used during development, and eventually extend our increased conceptual understanding to other animal models.
2. Dissecting the mechanism by which the transcriptional history of a cell influences its terminal fate
In every multicellular animal, specialized cells are produced from the zygote through coordinated cell divisions and progressive changes in gene expression. Cells follow such specific paths to reach their fate, and in C. elegans, these paths are invariant between individuals (Figure 1). At every cell division, cells face binary choices to go down one path or another. This allows us to identify interesting decision points during the production of a multicellular animal which we can probe at different levels.
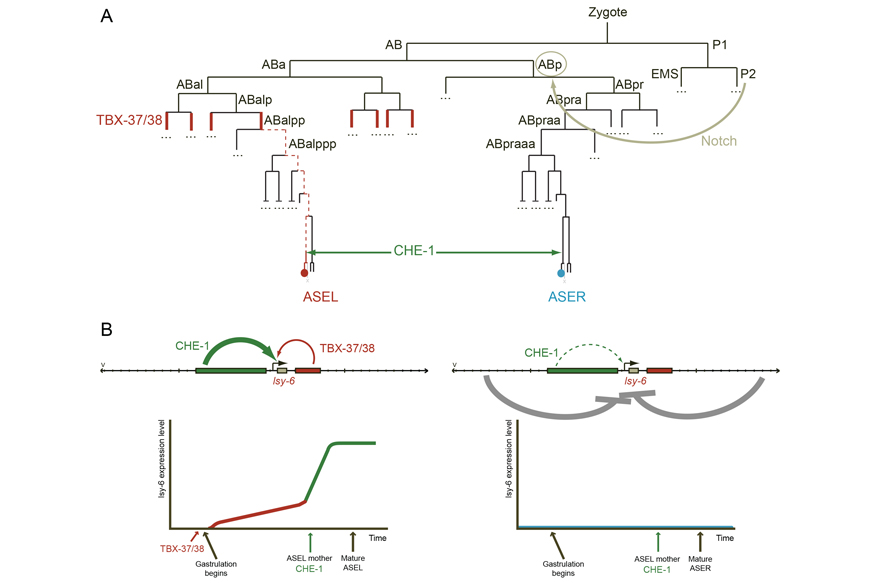
Knowing the developmental path of every cell has allowed us to identify a transcriptional mechanism for cell type diversification that we termed “prime and boost”. Through this mechanism, two cells that express the same terminal transcription factor and which should adopt the same fate are actually diversified because they originate from different lineages that differentially express another transcription factor early during their development. This early transcription factor, which we called the priming factor, is necessary for a specific gene locus to be activated by the terminal transcription factor five cell divisions later. Ultimately, the presence or absence of the priming factor early in development determines whether a specific locus is transcribed or not in the differentiated state and provides the basis for the production of two distinct cell identities (Figure 3). Our current efforts focus on trying to understand the mechanism by which transient expression of a transcription factor early in the history of a cell has an impact on the terminal fate of that cell. To achieve this, we are developing tools to gain access to genome-wide information of specific cell types in the developing C. elegans embryo and combining these with precise genetic manipulations.
Conceptually similar mechanisms have been proposed during the specification of different lineages in other animals. Our goal is to use the worm to dissect one such mechanism, test the extent to which such a strategy is used during development, and eventually extend our increased conceptual understanding to other animal models.
Selected Publications
- Alberti, C., Manzenreither, RA., Sowemimo, I., Burkard, TR., Wang, J., Mahofsky, K., Ameres, SL., Cochella, L. (2018). Cell-type specific sequencing of microRNAs from complex animal tissues. Nat Methods. 15(4):283-289.
- Alberti, C., Cochella, L. (2017). A framework for understanding the roles of miRNAs in animal development. Development. 144(14):2548-2559
- Drexel, T., Mahofsky, K., Latham, R., Zimmer, M., Cochella, L. (2016). Neuron type-specific miRNA represses two broadly expressed genes to modulate an avoidance behavior in C. elegans. Genes Dev.
- Cochella, L., Hobert, O. (2012). Diverse functions of microRNAs in nervous system development. Curr Top Dev Biol. 99:115-43
- Cochella, L., Hobert, O. (2012). Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell. 151(6):1229-42


