Finding and understanding cancer dependencies
Searching for exploitable genetic interactions in AML and PDAC
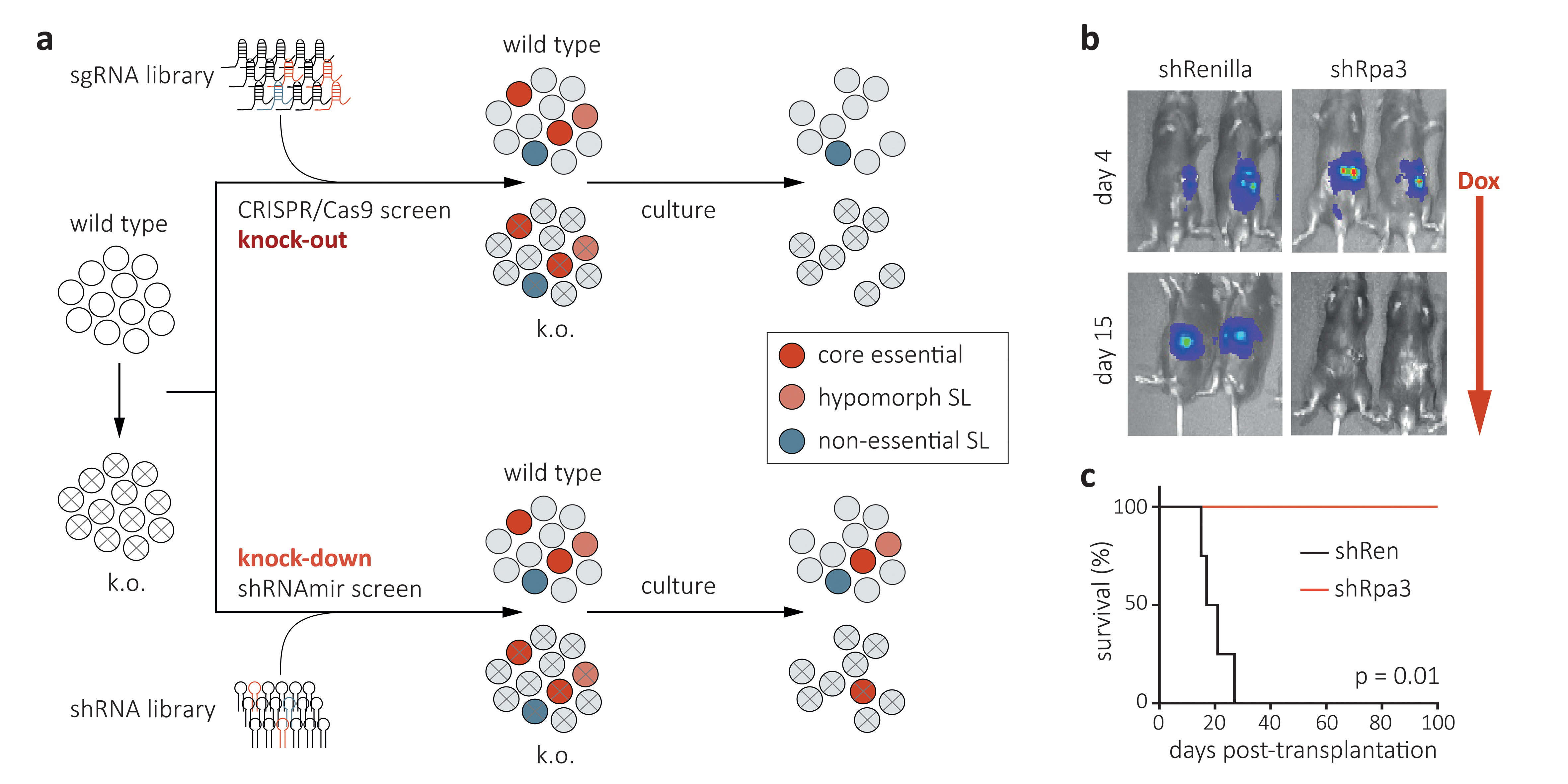
Cancer arises from complex and heterogeneous driver mutations that can affect over 500 cancer-associated genes. While most driver mutations are not directly druggable, synthetic-lethal interactions and the deregulation of fundamental cellular processes can result in cancer-specific dependencies that might be exploitable for therapy. The advent of loss-of-function genetic tools such as CRISPR/Cas9-, advanced RNAi-, and degron-technologies (reviewed in Housden & Muhar et al., Nature Reviews Genetics, 2017) has opened new frontiers in the search and study of such dependencies prior to drug development. Our lab develops and applies these tools for the identification and mechanistic study of exploitable dependencies in two major human cancers: acute myeloid leukemia (AML) and pancreatic adenocarcinoma (PDAC). Our experimental approach combines high-throughput CRISPR/Cas9- and shRNAmir-based genetic screens in defined murine and human cell lines (Fig. 1a) with loss-of-function studies in genetically engineered AML and PDAC mouse models (GEMMs), which provide an ideal system for validating and studying candidate therapeutic targets in a physiologic environment. Beyond stable and regulatable CRISPR/Cas9-based gene knock-out, we have developed advanced RNAi tools based on Tet-on regulatable miRNA-embedded shRNAs (shRNAmirs), which enable the in vivo suppression of candidate targets in established tumors, and thereby closely mimic a therapeutic intervention with a pharmacologic inhibitor (Fig. 1b). Beyond evaluating target suppression effects in cancer cells, we are employing Tet-on shRNAmir transgenic mice to genetically probe effects and possible toxicities in the microenvironment, the immune system and other normal tissues. Overall, our vision and mission are to develop and use cutting-edge functional genetic approaches to identify, validate and mechanistically study cancer-specific dependencies in AML and PDAC, and thereby guide the development of rational and effective cancer therapeutics.
Exploring and exploiting aberrant self-renewal in leukemia
In normal tissues, only stem cells possess the ability to self-renew, while all their progeny commits to a limited number of divisions before entering a terminal fate. Cancer cells lose this commitment to terminal differentiation and gain the ability to aberrantly self-renew (Fig. 2a). While genetic studies demonstrate that aberrant self-renewal can be reversed and therapeutically exploited, most associated mutations remain undruggable and common mechanisms underlying this phenomenon are poorly defined. Our lab explores these mechanisms and therapeutic targets for breaking aberrant self-renewal using three complementary approaches: (1) Using CRISPR/Cas9- and shRNAmir-based screens we systematically identify epigenetic regulators involved in maintaining aberrant cell states in genetically and histologically diverse leukemia subtypes (Fig. 2b). (2) Using reporter-based CRISPR/Cas9 screens we are deciphering regulators and co-factors of “undruggable” oncogenes and transcriptional master regulators, to understand the wiring of oncogenic regulatory networks and to explore new ways for therapeutically interfering with them. (3) By combining next generation RNAi or targeted protein degradation with advanced transcriptome and chromatin profiling methods, we are investigating primary regulatory functions of known and newly identified drivers of aberrant self-renewal.
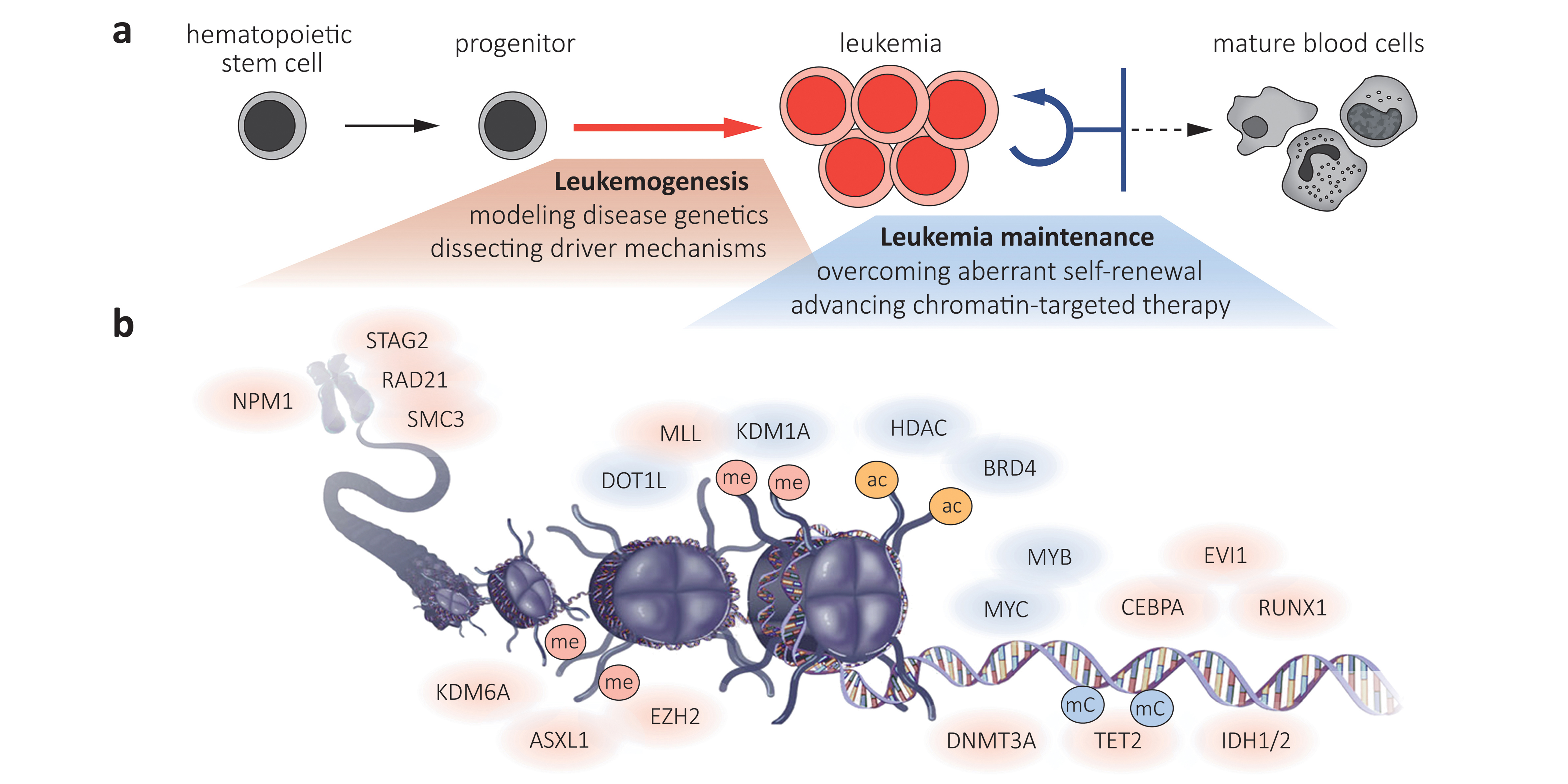
Illustration modified from Saygin and Carraway (Journal of Hematology & Oncology, 2017) under Creative Commons License 4.0 (http://creativecommons.org/licenses/by/4.0)
A prominent dependency that emerged from such screens is BRD4, a histone-acetyl reader whose partial suppression using RNAi or BET bromodomain inhibitors (BETi) triggers strong and selective therapeutic effects in a wide range of myeloid and B-cell leukemias. While BETi have already entered clinical trials less than three years after our initial study (Zuber & Shi et al., Nature, 2011), the mechanisms determining sensitivity and resistance to these agents remain incompletely understood. By integrating multiplexed shRNAmir screening and epigenetic profiling of sensitive and resistant cancers, we have identified compensatory gene regulatory pathways that promote primary and acquired resistance to BETi by restoring the transcription of critical target genes such as MYC (Rathert & Roth et al., Nature, 2015). Intriguingly, our and other studies indicate that such epigenetic ‘escape routes’ are hard-wired and predictable, suggesting that they can be exploited for the development of combinatorial or sequential targeted therapies, which is a major focus of ongoing projects.
Finding and probing immunomodulatory targets in tumor-associated T-cells
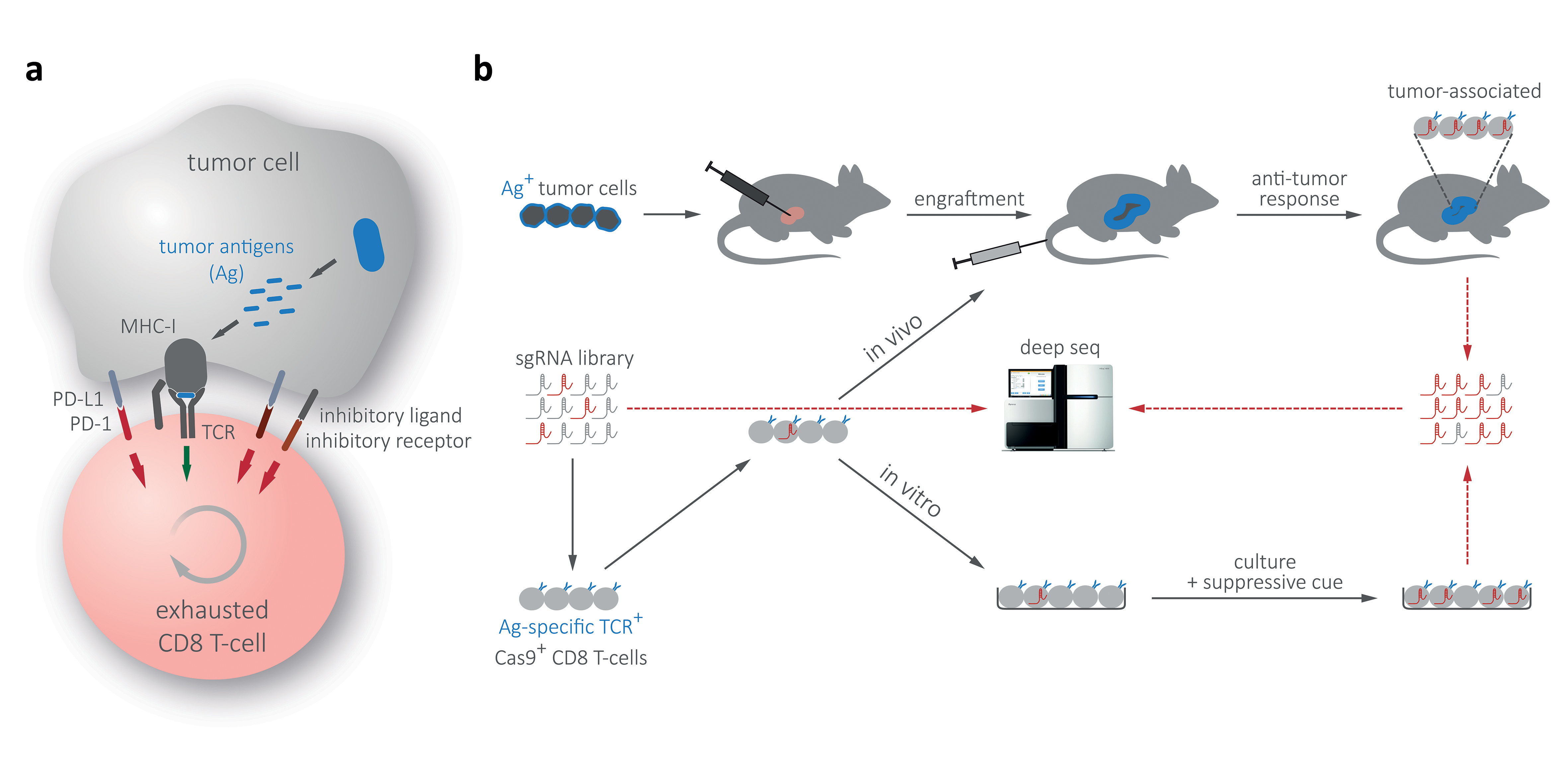
The groundbreaking success of checkpoint-blockade therapies in advanced melanoma is the single most impressive advance in clinical oncology. While this breakthrough showcases the potential of immunomodulatory therapies, clinical success of PD-1, PD-L1 and CTLA-4 blocking antibodies remains limited to certain cancer types, and key regulators and pathways involved in the establishment and maintenance of T-cell exhaustion are incompletely understood (Fig. 3a). To genetically study T-cell suppression pathways and search for additional (potentially small-molecule) targets, we have adapted RNAi- and CRISPR/Cas9-based screening methodology for multiplexed genetic screens in CD8 T-cells (Fig. 3b). Our experimental system is based on sequential transplantation of surrogate tumor antigen-expressing AML and PDAC models, followed by the adoptive transfer of tumor antigen specific CD8 T-cells that have been transduced in short-term culture with focused shRNAmir or CRISPR libraries. In these screens, shRNAs/sgRNAs targeting factors involved in T-cell suppression will promote the activity and proliferation of affected CD8 T-cells and, therefore, be positively selected within a complex pool, allowing their identification through next generation sequencing. In a complementary effort – addressing the quantitative limitations associated with in vivo screens – we expand the scale of our genetic screens to in vitro cultures. Following the establishment of surrogate tumor antigen positive GEMMs, suitable library vectors, CD8 transduction protocols and focused shRNA/sgRNA libraries, we have recently completed first multiplexed screens. Beyond validation studies in primary transplantation models and molecular characterization of the mode of T-cell suppression, we envision to probe validated targets in Tet-on shRNAmir transgenic mice, which provide a unique experimental strategy for modeling the effects of targeted therapies prior to drug development.
Selected Publications
-
Pechincha, C., Groessl, S., Kalis, R., de Almeida, M., Zanotti, A., Wittmann, M., Schneider, M., de Campos, RP., Rieser, S., Brandstetter, M., Schleiffer, A., Müller-Decker, K., Helm, D., Jabs, S., Haselbach, D., Lemberg, MK., Zuber, J., Palm, W. (2022) Lysosomal enzyme trafficking factor LYSET enables nutritional usage of extracellular proteins. Science. 378(6615):eabn5637
-
de Almeida, M., Hinterndorfer, M., Brunner, H., Grishkovskaya, I., Singh, K., Schleiffer, A., Jude, J., Deswal, S., Kalis, R., Vunjak, M., Lendl, T., Imre, R., Roitinger, E., Neumann, T., Kandolf, S., Schutzbier, M., Mechtler, K., Versteeg, GA., Haselbach, D., Zuber, J. (2021) AKIRIN2 controls the nuclear import of proteasomes in vertebrates. Nature. 599(7885):491-496
-
Muhar, M., Ebert, A., Neumann, T., Umkehrer, C., Jude, J., Wieshofer, C., Rescheneder, P., Lipp, J.J., Herzog, V.A., Reichholf, B., Cisneros, D.A., Hoffmann, T., Schlapansky, M.F., Bhat, P., von Haeseler, A., Köcher, T., Obenauf, A.C., Popow, J., Ameres, S.L., and Zuber, J. (2018). SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science (DOI: 10.1126/science.aao2793)
-
Rathert, P., Roth, M., Neumann, T., Muerdter, F., Roe, JS., Muhar, M., Deswal, S., Cerny-Reiterer, S., Peter, B., Jude, J., Hoffmann, T., Boryń, ŁM., Axelsson, E., Schweifer, N., Tontsch-Grunt, U., Dow, LE., Gianni, D., Pearson, M., Valent, P., Stark, A., Kraut, N., Vakoc, CR., Zuber, J. (2015). Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 525(7570):543-7
- Zuber, J., Shi, J., Wang, E., Rappaport, AR., Herrmann, H., Sison, EA., Magoon, D., Qi, J., Blatt, K., Wunderlich, M., Taylor, MJ., Johns, C., Chicas, A., Mulloy, JC., Kogan, SC., Brown, P., Valent, P., Bradner, JE., Lowe, SW., Vakoc, CR. (2011). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 478(7370):524-8
Join us
- Master students and Post-docs: Contact Johannes Zuber with a letter of intent detailing why you want to join the lab.
- PhD students: Calls open 1 March and 1 September, apply here:
Vienna BioCenter PhD Program




