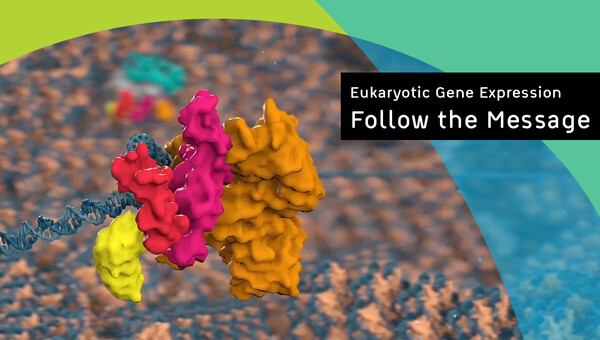
Eukaryotic gene expression begins with mRNA production in the nucleus. The newly made precursor mRNA (pre-mRNA) undergoes multiple maturation steps, which are critical for producing fully mature mRNA. The nuclear mRNA must then be recognized as mature and exported to cytoplasm, where it serves as a blueprint for translation. Throughout its life cycle, mRNA is never left exposed but is instead bound by many proteins and complexes that exchange dynamically. These proteins facilitate the separate maturation steps themselves, coordinate between them, and directly mark the mRNA upon successful completion of maturation. Our lab aims to understand these molecular mechanisms of gene expression. Currently, we are fascinated to understand how nuclear mRNA is matured, how the completion of maturation is sensed, and how the mature mRNA is ultimately exported to the cytoplasm.
Figure 1: Cryo-EM structure of the human core transcription and export complex. For details see Pühringer*, Hohmann* et al., eLife (2020) 9, e61503.
A key mRNA maturation event is the splicing of the pre-mRNA. Splicing is carried out by the spliceosome, a dynamic multi-megadalton machine that excises non-coding introns from the pre-mRNA. The splicing machinery assembles anew on each intron through the step-wise addition of five U-rich small nuclear RNA–protein complexes (U1, U2, U4, U5, U6), and several accessory factors, comprising over 100 proteins in humans. During spliceosome assembly, alternative exons can be selected, leading to distinct mRNA isoforms from a single gene that can encode proteins with dramatically different functions. Splicing coordinates with downstream events, such as the packaging and export of mRNA. Since mRNA from different genes vary in their length and sequence, mRNA packaging and export needs to be both highly selective for mature mRNA but at the same time highly permissive to the great diversity of mRNAs made by our genome. Despite the important roles of these maturation steps in gene expression, many questions regarding their mechanisms remain. What is the structural and mechanistic basis of splicing regulation? How are maturation events such as splicing packaging physically coordinated? How does a limited set of proteins allow for the maturation of the vast diversity of mRNAs? Finally, how are defects in the mRNA life cycle linked to human disease?
Figure 2: Cryo-EM structure of the spliceosome B complex. The structure contains the pre-precursor mRNA substrate (pre-mRNA, black), 52 spliceosome proteins, and four small nuclear RNAs that are coloured according to ribonucleoprotein complex identity (U2, green; U4, yellow; U5, blue; U6, red). For details see Plaschka et al., Nature (2017) 546, 617-621.
To shed light on these questions, we determine the three-dimensional structures of complexes involved in human mRNA maturation and study them in cells using next-generation sequencing and advanced microscopy methods. We prepare mRNA-protein complexes from endogenous or recombinant sources and use cryo-electron microscopy (cryo-EM) and X-ray crystallography to resolve their structures. Complementary methods, such as protein crosslinking and mass spectrometry, facilitate an integrative modelling of the complexes. We additionally use functional biochemistry, RNA sequencing, and advanced microscopy methods to further elucidate the molecular mechanisms in vitro and in vivo. Through these approaches we have previously studied mechanisms of RNA polymerase II transcription, pre-mRNA splicing, and, most recently, mRNA nuclear export (Fig. 1, 2).
Open positions: The Plaschka Lab welcomes applications for Master, PhD, and Postdoc positions, who wish to apply state-of-the-art structural biology, sequencing, and advanced microscopy methods to understand the complex process of mRNA regulation.
Clemens Plaschka lab videos
Selected Publications
-
Vorländer, M. K., Rothe, P., Kleifeld, J., Cormack, E., Veleti, L., Riabov-Bassat, D., Fin, L., Phillips, A. W., Cochella, L., Plaschka, C. (2024): Mechanism for the initiation of spliceosome disassembly. Nature.
-
Pacheco-Fiallos, B., Vorländer, MK., Riabov-Bassat, D., Fin, L., O'Reilly, FJ., Ayala, FI., Schellhaas, U., Rappsilber, J., Plaschka, C. (2023) mRNA recognition and packaging by the human transcription-export complex. Nature.
-
T. Pühringer*, U. Hohmann*, L. Fin, B. Pacheco-Fiallos, U. Schellhaas, J. Brennecke, C. Plaschka. Structure of the human core transcription-export complex reveals a hub for multivalent interactions. eLife (2020) 9, e61503. *Equal contribution.
-
C. Plaschka*#, P.-C. Lin*#, C. Charenton, K. Nagai#. Prespliceosome structure provides insight into spliceosome assembly and regulation. Nature (2018) 558, 419-422. *Equal contribution. #Co-corresponding authors.
- C. Plaschka*#, P.-C. Lin*#, K. Nagai#. Structure of a pre-catalytic spliceosome. Nature (2017) 546, 617-621. *Equal contribution. #Co-corresponding authors.
Join us
- Master students and Post-docs: Contact Clemens Plaschka with a letter of intent detailing why you want to join the lab.
- PhD students: Calls open 1 March and 1 September, apply here:
Vienna BioCenter PhD Program