Molecular mechanisms of antibody diversification
Research activities
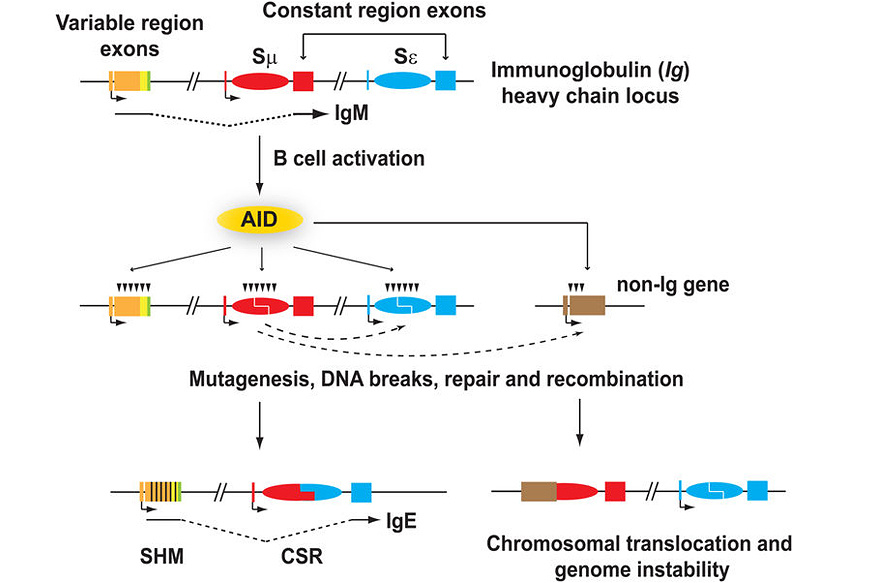
Antibody diversification occurs at the immunoglobulin (Ig) loci and consists of two processes: somatic hypermutation (SHM) which modulates the antigen-binding properties of the antibody, and class switch recombination (CSR) which generates various antibody isotypes (Figure 1). Antibody maturation is triggered by activation-induced deaminase (AID), which generates mismatches in the Ig DNA sequence by converting cytosines into uracils on single-stranded DNA. AID can also target non-Ig genes, such as the oncogenes, c-myc and Bcl6, albeit to a much lesser extent (Figure 1). These lesions are then processed by various means, involving error-prone repair, double strand breaks (DSBs), and non-homologous end-joining (NHEJ), resulting in SHM and CSR (Figure 1). AID-induced DSBs at non-Ig genes like c-myc may also lead to c-myc/IgH translocations and lymphoid malignancies (Figure 1). In our lab, we are interested in understanding the role of transcription and co-transcriptional factors involved in AID targeting and mutagenesis, and we determine how AID targeting is restricted to specific loci in B cells.
Transcriptional regulation of AID targeting
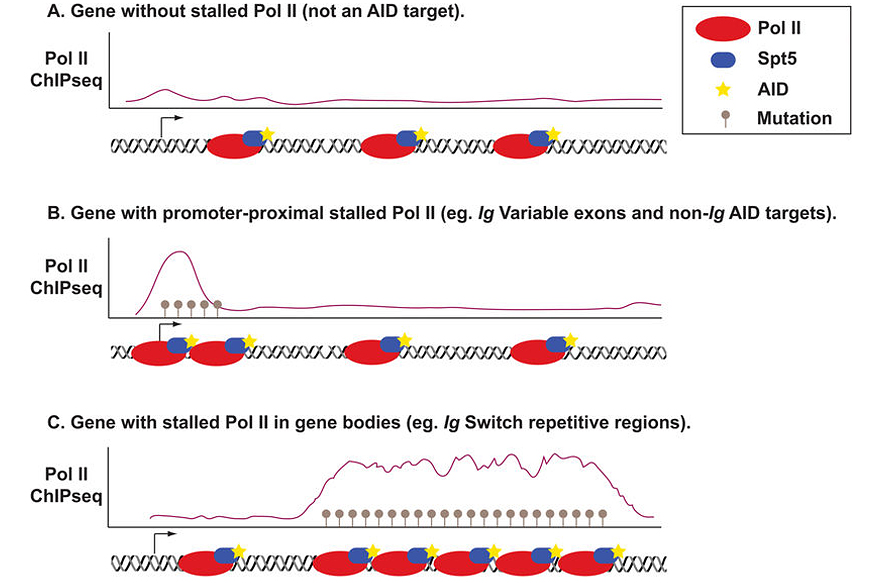
We have shown that the RNA polymerase II (Pol II) stalling and elongation factor, Spt5, is required for CSR and AID targeting. Based on our findings we proposed a model for AID targeting via Spt5 and Pol II stalling (Figure 2), wherein AID is preferentially targeted to regions of Pol II stalling through direct interaction with Spt5. Stalling provides AID with long residence times at, and access to, its ssDNA substrate within the transcription bubble. In this manner, mutations are focused on specific genomic regions which coincide with the known mutation profiles of AID as well as hotspots of AID-dependent translocations (Figure 2).
We are currently developing tools and genetic models to further study the impact of Pol II stalling and Spt5 function in diversifying B cells. Our preliminary findings suggest entirely new modes of gene regulation by Spt5 during B cell activation. We are currently investigating how this influences AID regulation, Ig gene expression, and AID targeting during CSR and SHM.
Cell cycle regulation of antibody diversification
Antibody diversification, both in vivo and in vitro, occurs in highly proliferating B cells. In vitro studies clearly show a correlation between the magnitude of CSR and cell division. The reasons for this are unclear. Cell cycle regulation and/or DNA replication have not been implicated in CSR and SHM so far. Using RNAi screening in B cells, we disclosed the role of cell cycle and DNA replication factors in CSR. Preliminary findings show that these factors associate with AID and Spt5, both in vitro and in vivo, and are localized to AID target genes on a genome-wide basis. We are currently investigating the mechanisms by which these factors regulate CSR, and how CSR is coordinated during the cell cycle in proliferating and hypermutating B cells.
Identification and characterization of novel factors in CSR and SHM
To gain insights into the mechanism of CSR, we performed an RNAi screen for the purpose of identifying new factors involved in this process. We are currently investigating interesting candidates determined from this screen, using mouse genetics, biochemical analyses, and genomics. In addition, we are developing new assays to quantitatively test for SHM in B cells, which will allow us to perform RNAi studies to address the precise role of various putative factors involved in SHM and identify new players in this process.
Our long-term goal is to determine how AID targeting is tightly restricted to specific regions within the transcribed Ig locus, especially during SHM. This enigmatic aspect of the phenomenon eludes a clear explanation, but is critical for the immune response.
Selected Publications
- Costea, J., Schoeberl, U. E., Malzl, D., Von der Linde, M., Fitz, J., Makharova, M., Goloborodko, A., Pavri, R. (2023) A de novo transcription-dependent TAD boundary underpins critical multiway interactions during antibody class switch recombination. Mol Cell; S1097-2765(23)00037-0
- Peycheva, M., Neumann, T., Malzl, D., Nazarova, M., Schoeberl, U. and Pavri, R. (2022) DNA replication timing directly regulates the frequency of oncogenic chromosomal translocations. Science 377, 6612
- Fitz, J., Neumann, T., Steininger, M., Wiedemann, EM., Garcia, AC., Athanasiadis, A., Schoeberl, UE., Pavri, R. (2020). Spt5-mediated enhancer transcription directly couples enhancer activation with physical promoter interaction. Nature Genetics 52, 505–515
- Fitz, J., Neumann, T., Pavri, R. (2018). Regulation of RNA polymerase II processivity by Spt5 is restricted to a narrow window during elongation. EMBO J. 37(8)
- Wiedemann, EM., Peycheva, M., Pavri, R. (2016). DNA Replication Origins in Immunoglobulin Switch Regions Regulate Class Switch Recombination in an R-Loop-Dependent Manner. Cell Rep. 17(11):2927-2942
Join us
- Master students and Post-docs: Contact Rushad Pavri with a letter of intent detailing why you want to join the lab.
- PhD students: Calls open 1 March and 1 September, apply here:
Vienna BioCenter PhD Program
How DNA replication timing regulates chromosomal translocations
We use a YouTube plugin to display social media content on this page, which places requests to YouTube servers. These requests make your IP address visible to YouTube, who may use it in accordance with their data privacy policy. Please agree to make the YouTube video visible. You can find more information in our privacy settings.
Accept