Functional genomics of the innate immune system
While canonical immune sensing relies on pattern-recognition of microbial components, the innate immune system can also recognize the virulence-associated enzymatic activities of pathogens. To avoid immune defences and exploit host cells, pathogens produce virulence factors that disrupt critical immune pathways. As postulated by the guard hypothesis, immunity in plants relies on a secondary line of pathogen detection, which monitors (or ‘guards’) the integrity of host immune defines pathways.
Virulence factor-mediated disruption of a guarded pathway triggers activation of a secondary guard immune response. While this guard-type immunity is a common feature of plant immune systems, the overall importance of guard-immunity in mammals is less clear. We have recently identified “self-guarding” as a new principle for detection of virulence factors in which the immune system senses “perturbed-self” as opposed to “non-self”. Self-guarding relies on a bi-functional protein that combines guarded function and guard function into a single polypeptide.
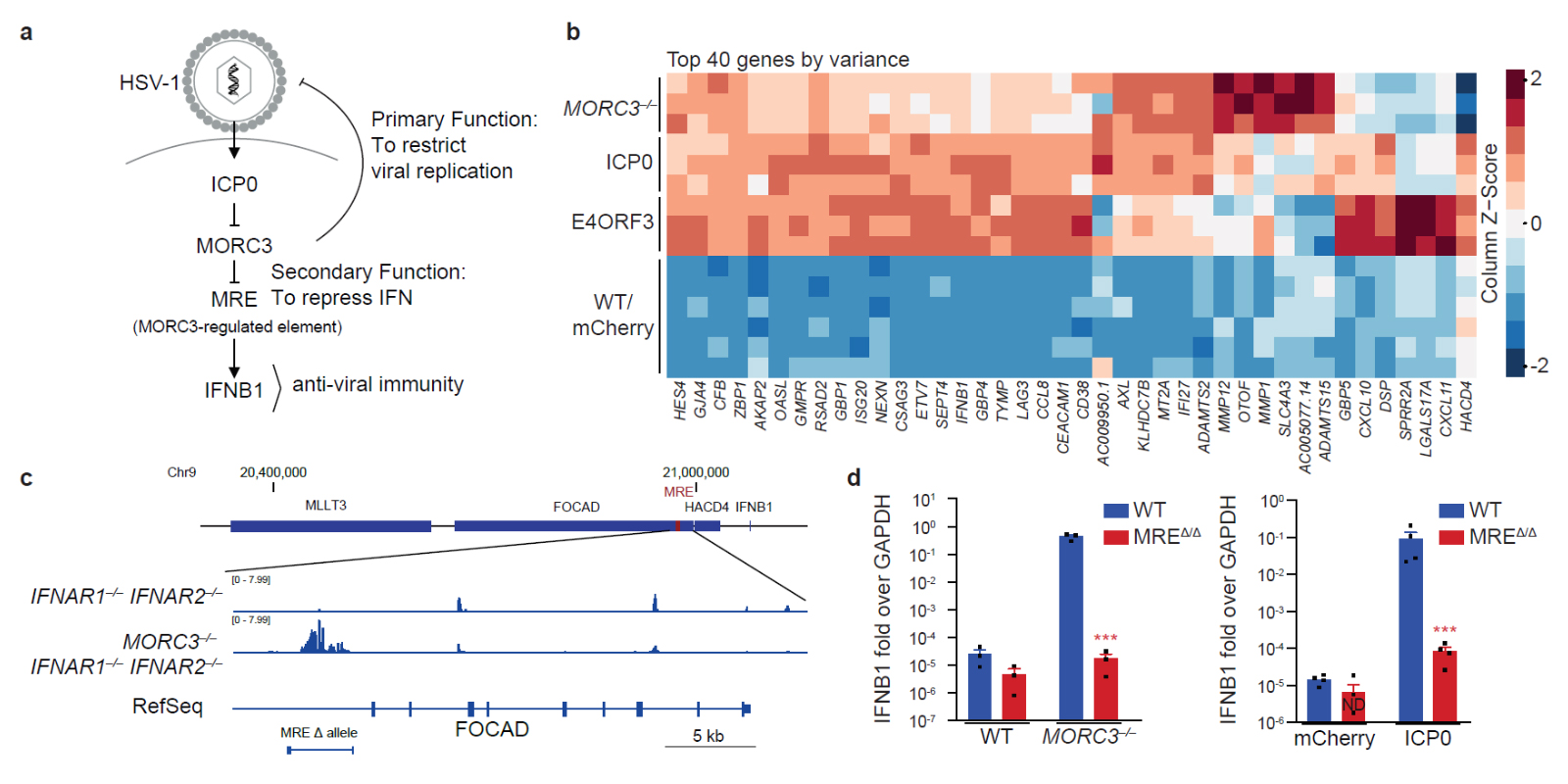
(a) The self-guarded MORC3 pathway is attacked by ICP0 from HSV-1 to alleviate its primary direct anti-viral function. MORC3 has a secondary function to repress the MRE-DNA element. If MORC3 is degraded by ICP0, the MRE activates anti-viral interferon.
(b) Genetic activation (in MORC3–/– monocytes) and ICP0-mediated activation of the MORC3 pathway induce a similar transcriptional program.
(c) ATAC-seq identifies the MORC3-regulated element (MRE) that is repressed by MORC3.
(d) The MRE senses perturbation of MORC3 by genetic tools or viral ICP0 and induces IFN.
Our first example is the bi-functional protein MORC3, a direct anti-viral restriction factor that represses replication of viruses like Herpes Simplex Virus-1 (HSV-1). Consequently, HSV-1 employs the virulence factor ICP0 to attack and degrade MORC3 to restore viral replication. To sense this viral attack, MORC3 has a secondary function as a repressor of a novel regulatory DNA-element that we call the MRE (MORC3-regulated element). In response to virus-mediated degradation of MORC3, the MRE drives activation of the anti-viral cytokine Type I interferon (IFN) by activating the IFNB1 gene in cis. Virulence factor-mediated attack of MORC3 thus inadvertently de-represses the IFNB1 locus.
The cytokine IFNB1 then drives anti-viral defences by alerting adjacent cells to the presence of a virus. MORC3’s bi-functionality forces the pathogen to “pick its poison”: either the pathogen is restricted by its primary anti-viral activity or, if the pathogen interferes with this activity, a secondary restrictive anti-viral IFN response is unleashed. The MORC3 pathway is an elegant example of reciprocal countermeasures in the arms races between pathogens and their hosts.
We use functional genomics to identify genetic components of host-pathogen interactions, innate immune sensing, and immune homeostasis. This involves forward and reverse genetics in human cells, mice, and model pathogens. One focus lies on the BLaER1 cell culture system of human monocytes that we have pioneered for genetically dissecting human innate immune pathways. The lab’s overall interests centre around these topics:
Innate immune sensing: How are pathogens recognized by the innate immune system? Which receptors directly recognize microbes as non-self? Which sensors indirectly report microbial presence by sensing perturbed-self? Which microbial components/virulence factors are sensed?
Innate immune signalling: What are the functional consequences of immune receptor activation? How are immune genes activated and what are the regulatory genetic circuits?
Innate immune therapies: Can we utilize our insights into the innate immune system to develop novel therapies to enhance inflammation (against cancer) or dampen inflammation (in sterile inflammatory conditions)?
Join the lab
- Master students and Post-docs: Contact the Moritz Gaidt with a letter of intent detailing why you want to join the lab.
- PhD students: Calls open 1 March and 1 September, apply here:
Vienna BioCenter PhD Program
Selected Publications
- M.M. Gaidt#, A. Morrow, M.R. Fairgrieve, J.P. Karr, N. Yosef and R.E. Vance#, Self-guarding of MORC3 enables virulence factor-triggered immunity, Nature. In press (2021). #co-corresponding authors
- M.M. Gaidt, T.S. Ebert, D. Chauhan, K. Ramshorn, F. Pinci, S. Zuber, F. O’Duill, J.L. Schmid-Burgk, F. Hoss, R. Buhmann, G. Wittmann, E. Latz, M. Subklewe, V. Hornung, The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3, Cell. 171 (2017) 1110-1124.e18.
- M.M. Gaidt, T.S. Ebert, D. Chauhan, T. Schmidt, J.L. Schmid-Burgk, F. Rapino, A.A.B. Robertson, M.A. Cooper, T. Graf, V. Hornung, Human Monocytes Engage an Alternative Inflammasome Pathway, Immunity. 44 (2016) 833–846.
- M.M. Gaidt, F. Rapino, T. Graf, V. Hornung, Modeling primary human monocytes with the trans–differentiation cell line BLaER1, in: Methods Mol. Biol., 2018: pp. 57–66.
Rresearch questions and approaches
We use a YouTube plugin to display social media content on this page, which places requests to YouTube servers. These requests make your IP address visible to YouTube, who may use it in accordance with their data privacy policy. Please agree to make the YouTube video visible. You can find more information in our privacy settings.
Accept