David Keays is an Adjunct Investigator at the IMP and Professor at the Ludwig-Maximilians-Universität München. You find more detailed information about his work and research group at: https://www.neuro.bio.lmu.de/research_groups/res-keays_d/
Microtubules and Magnetoreception
The driving philosophy of my laboratory is to unravel nature’s wonder by relentless attention to detail. We are interested in two questions in neuroscience:
(1) How do animals sense the Earth’s magnetic field?
(2) What role does the microtubule cytoskeleton play in the developing brain?
In tackling these two questions we adopt an inter-disciplinary approach that exploits a broad range of genetic, histological, imaging, cellular and molecular tools. Moreover, we embrace the same rigorous reductionist mindset that aims to perform carefully controlled experiments, the results of which are critically analysed.
Magnetoreception
Using maps, compasses, and sextants, mariners in the early 1500's developed the first methods to navigate the open sea; heralding an age of exploration as humanity set sail for the horizon. Yet long before this time evolution had equipped life on the planet with a biological global positioning system that was far superior to those early navigational tools – the Magnetic Sense. While there is unequivocal behavioural evidence demonstrating that this faculty exists, it is the least understood of all senses. The location of the primary sensors, the underlying biophysical mechanisms, and the neurological basis of the sense are unknown. Currently, there are three ideas that aim to explain how magnetosensation might work: (1) a light sensitive radical pair based model; (2) magnetite based magnetoreception; and (3) electromagnetic induction. Previous studies have asserted that pigeons employ a magnetite containing sensory apparatus located at six specific loci in the subepidermis of the beak. We have shown that this dogma is false, and in reality clusters of iron-rich cells in the beak of pigeons are macrophages not magnetosensitive neurons (Treiber et al, Nature 2012). It is apparent that the true magnetosensors remain to be found.
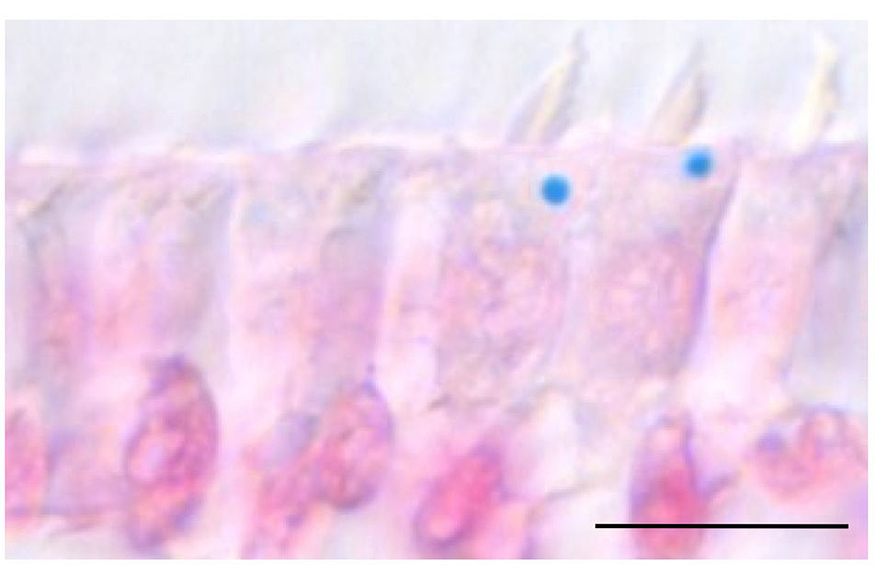
Our goal is to identify the molecules, cells and circuits that underlie the magnetic sense in pigeons. To achieve this objective we employ an assay that assesses neuronal activation within the pigeon brain, following exposure to magnetic fields generated by Helmholtz coils. These experiments have implicated the vestibular nuclei and the inner ear of pigeons in the magnetic sense, resulting in the discovery of an iron-rich organelle, the “cuticulosome” which resides in avian hair cells (Fig.1. Lauwers et al, Current Biology 2013).
Current Research Projects
1. Where are the primary magnetosensors? Having established a physiologically relevant readout for the magnetic sense, we can now ask where do the primary magnetosensors reside. To do so we are employing anatomical perturbations (e.g. hair cell ablation with antibiotics), which we expect will be superseded by genetic methods. We are complementing this work with a systematic search for magnetite within the pigeon inner ear using synchrotron radiation and an elemental characterisation of the cuticulosome.
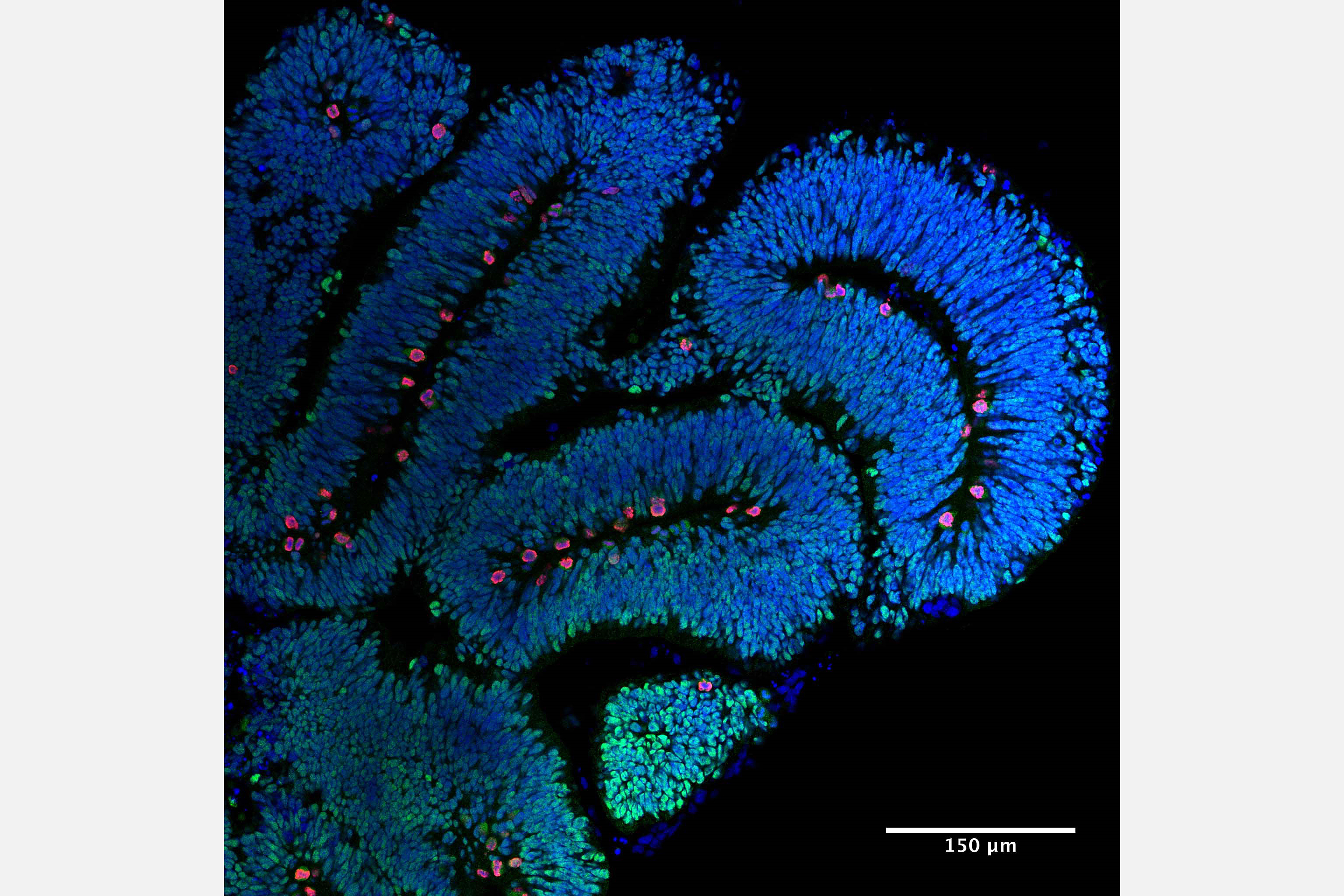
2. What neuronal circuits process magnetic information? To gain further insight into the underlying circuitry that processes magnetic information we have established an iDISCO clearing protocol for the pigeon brain that results in a translucent brain that can be stained with markers such as c-fos followed by light sheet microscopy (Fig 2.). We are employing this technology to identify the neuronal circuits that process magnetic information and to interrogate the underlying biophysical mechanisms.
3. How is magnetic information encoded in the avian brain? To address this question brain we have built a 2-photon microscope that permits in vivo calcium imaging while exposing pigeons to precise magnetic stimuli. Coupled with a genetically encoded calcium indicator (e.g GCaMP6) delivered by an adeno associated virus we are able to study which components of the magnetic field elicit neuronal activity (i.e. intensity, polarity and inclination) and how this information is integrated into existing neuronal networks.

Microtubules and Neurodevelopment
The construction of the vertebrate brain is dependent on a complex cascade of biological processes that include mitotic division, relocation of migrating neurons, and the extension of dendritic and axonal processes. To gain insight into the molecules that mediate these cellular events my laboratory has relied on unbiased phenotypic screens in mice, the generation of novel transgenic mouse mutants, coupled with the study of human neurodevelopmental disorders. This work has highlighted the importance of the microtubule cytoskeleton and associated proteins in the construction of the human brain.
We have shown that mutations in the -tubulin gene TUBA1A cause lissencephaly a neurodevelopmental disease characterised by a smooth cortex, epilepsy and severe intellectual impairment. The tubulins are a multi-gene family that encode for the constituents of microtubules; in humans there are 8 α- and 9 β-tubulin genes. Since our discovery that mutations in TUBA1A cause lissencephaly a host of neurological disease states have been associated with the tubulins. For instance, it is now known that variants in TUBB3 cause defects in axon guidance, mutations in TUBA4A cause motor neuron disease, and mutations in the β -tubulin gene TUBB5 cause microcephaly. We have built on this work further demonstrating that mutations in the microtubule associated protein EB2 cause the unusual “Michelin tyre” syndrome, and that variants in a microtubule associated kinase (MAST1) cause a rare disorder characterised by an enlarged corpus callosum. We are interested in understanding how these mutations cause a spectrum of neurodevelopmental diseases and what role these genes play in the developing brain.
Current Research Projects
1. Do tubulin genes have specific functions? An emerging theme from our work is that mutations in different tubulin genes give rise to overlapping but distinct disorders. It has been further noted that each tubulin protein has a unique variant C-terminal region and is expressed differentially. These observations support the idea that individual tubulin genes encode functionally distinct polypeptides. Known as the multi-tubulin hypothesis this idea was first proposed more than 40 years ago by Fulton and Simpson, but has not been rigorously testing in a vertebrate system. We are exploiting the power of mouse genetics, coupled with the CRISPR-cas9 genome editing system to test this hypothesis, defining the role of different tubulin genes in the developing brain.
2. Organoids and tubulin gene mutations. We are interested in understanding how mutations in the tubulin genes cause neurodevelopmental disease. To gain mechanistic insight into this question in a human context we are exploiting the power of human cerebral organoids to model the tubulinopathies. Drawing on our established network of clinicians, we generate iPSCs, which are then employed to generate “mini brains”. We carefully analyse these organoids alongside controls that have been subject to genetic repair using the CRISP-cas9 genome editing system. We use a variety of methods including brain clearing (iDISCO), light sheet microscopy, live cell imaging, and transcriptomic methods to gain insight into the underlying molecular pathology.
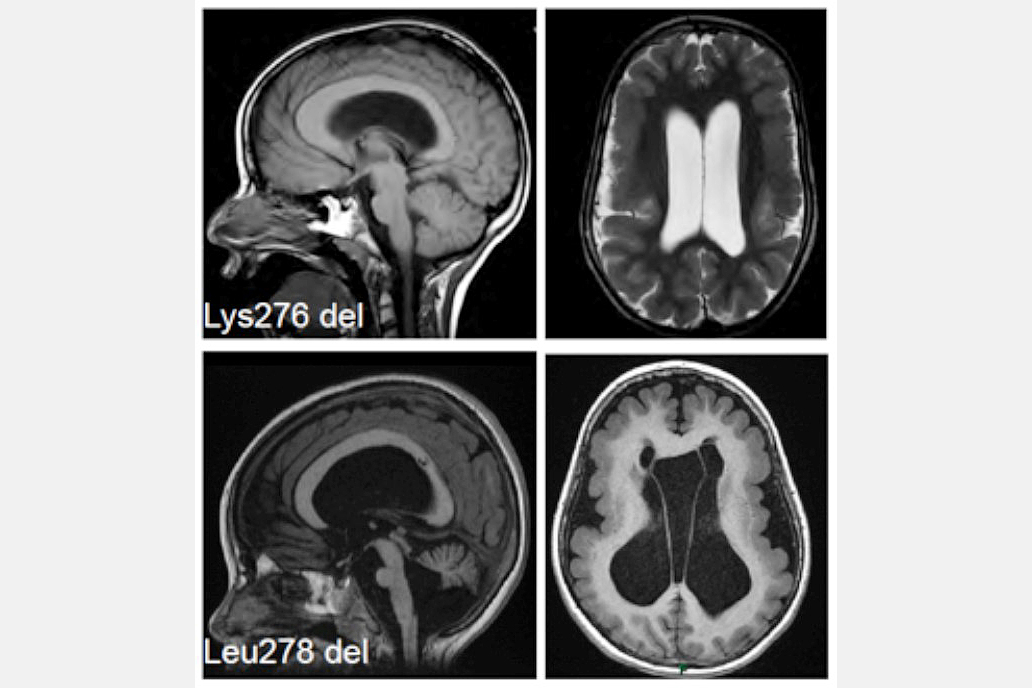
3. What role does the MAST family play in the developing brain? We have recently shown that microdeletions in MAST1 cause a rare disease characterised by a "mega" corpus callosum, and implicated this gene in autism spectrum disorder (Fig. 4). There are four MAST family proteins (MAST1-4), we are largely uncharacterised. Our goal is to define their expression pattern, identify their phosphorylation targets, understand how they regulate microtubule function, and explore the effects of their ablation. To achieve these goals and understand how mutations in the MAST family cause neurological disease we have generated a variety of novel mouse models that recapitulate patient mutations, and reprogrammed iPSCs for organoid development.
Collaborators
Professor Nick Cowan (NYU, New York) – Tubulin biochemistry
Professor Jamel Chelly (Inserm, Paris) – Tubulin and lisssencephaly
Professor Jonathan Flint (Oxford, UK) – Neurodevelopmental mouse mutants
Selected Publications
- Gstrein, T., Edwards, A., Přistoupilová, A., Leca, I., Breuss, M., Pilat-Carotta, S., Hansen, AH., Tripathy, R., Traunbauer, AK., Hochstoeger, T., Rosoklija, G., Repic, M., Landler, L., Stránecký, V., Dürnberger, G., Keane, TM., Zuber, J., Adams, DJ., Flint, J., Honzik, T., Gut, M., Beltran, S., Mechtler, K., Sherr, E., Kmoch, S., Gut, I., Keays, DA. (2018). Mutations in Vps15 perturb neuronal migration in mice and are associated with neurodevelopmental disease in humans.Nat Neurosci. 21(2):207-217
- Edelman, NB., Fritz, T., Nimpf, S., Pichler, P., Lauwers, M., Hickman, RW., Papadaki-Anastasopoulou, A., Ushakova, L., Heuser, T., Resch, GP., Saunders, M., Shaw, JA., Keays, DA. (2015). No evidence for intracellular magnetite in putative vertebrate magnetoreceptors identified by magnetic screening. Proc Natl Acad Sci U S A. 112(1):262-7
- Lauwers, M., Pichler, P., Edelman, NB., Resch, GP., Ushakova, L., Salzer, MC., Heyers, D., Saunders, M., Shaw, J., Keays, DA. (2013). An iron-rich organelle in the cuticular plate of avian hair cells. Curr Biol. 23(10):924-9
- Treiber, CD., Salzer, MC., Riegler, J., Edelman, N., Sugar, C., Breuss, M., Pichler, P., Cadiou, H., Saunders, M., Lythgoe, M., Shaw, J., Keays, DA. (2012). Clusters of iron-rich cells in the upper beak of pigeons are macrophages not magnetosensitive neurons. Nature. 484(7394):367-70
- Keays, DA., Tian, G., Poirier, K., Huang, GJ., Siebold, C., Cleak, J., Oliver, PL., Fray, M., Harvey, RJ., Molnár, Z., Piñon, MC., Dear, N., Valdar, W., Brown, SD., Davies, KE., Rawlins, JN., Cowan, NJ., Nolan, P., Chelly, J., Flint, J. (2007). Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 128(1):45-57
Grants


